Biomedical Computing
Biomedical computing combines the diagnostic and investigative aspects of biology and medical science with the power and problem-solving capabilities of modern computing. Computers are used to accelerate research learning, simulate patient behavior and visualize complex biological models.
Jeff Weiss
Computational Biomechanics
Orly Alter
Computational Biology
Tamara Bidone
Computational Models
Simulations of Biological Systems
Multi-Physics Models of Cancer Cells
Centers and Labs:
- Center for Integrative Biomedical Computing
- Muskuloskeletal Research Laboratory
- Genomic Signal Processing Lab
- Computational Biomechanics Group
Funded Research Projects:
Publications in Biomedical Computing:
  Extensions to a manifold learning framework for time-series analysis on dynamic manifolds in bioelectric signals B. Erem, R.M. Orellana, D.E. Hyde, J.M. Peters, F.H. Duffy, P. Stovicek, S.K. Warfield, R.S. MacLeod, G. Tadmor, D.H. Brooks. In Physical Review E, Vol. 93, No. 4, American Physical Society, apr, 2016. DOI: 10.1103/physreve.93.042218 This paper addresses the challenge of extracting meaningful information from measured bioelectric signals generated by complex, large scale physiological systems such as the brain or the heart. We focus on a combination of the well-known Laplacian eigenmaps machine learning approach with dynamical systems ideas to analyze emergent dynamic behaviors. The method reconstructs the abstract dynamical system phase-space geometry of the embedded measurements and tracks changes in physiological conditions or activities through changes in that geometry. It is geared to extract information from the joint behavior of time traces obtained from large sensor arrays, such as those used in multiple-electrode ECG and EEG, and explore the geometrical structure of the low dimensional embedding of moving time windows of those joint snapshots. Our main contribution is a method for mapping vectors from the phase space to the data domain. We present cases to evaluate the methods, including a synthetic example using the chaotic Lorenz system, several sets of cardiac measurements from both canine and human hearts, and measurements from a human brain. |
  Optimization of focality and direction in dense electrode array transcranial direct currentstimulation (tDCS) S. Guler, M. Dannhauer, B. Erem, R.S. Macleod, D. Tucker, S. Turovets, P. Luu, D. Erdogmus, D. Brooks. In Journal of Neural Engineering, Vol. 13, No. 3, IOP Publishing, pp. 036020. May, 2016. DOI: 10.1088/1741-2560/13/3/036020 OBJECTIVE: |
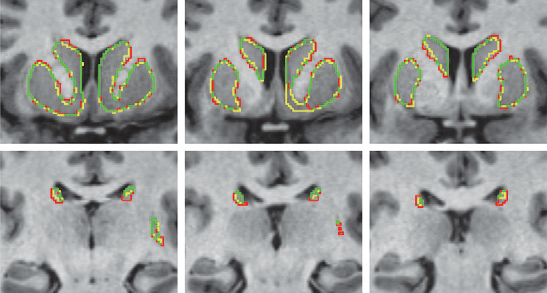
  Quantitative comparison of cortical bone thickness using correspondence-based shape modeling in patients with cam femoroacetabular impingement P.R. Atkins, S.Y. Elhabian, P. Agrawal, M.D. Harris, R.T. Whitaker, J.A. Weiss, C.L. Peters, A.E. Anderson. In Journal of Orthopaedic Research, Wiley-Blackwell, Nov, 2016. DOI: 10.1002/jor.23468 The proximal femur is abnormally shaped in patients with cam-type femoroacetabular impingement (FAI). Impingement |
  The role of blood vessels in high-resolution volume conductor head modeling of EEG L.D.J. Fiederer, J. Vorwerk, F. Lucka, M. Dannhauer, S. Yang, M. Dümpelmann, A. Schulze-Bonhage, A. Aertsen, O. Speck, C.H. Wolters, T. Ball. In NeuroImage, Vol. 128, Elsevier, pp. 193--208. March, 2016. DOI: 10.1016/j.neuroimage.2015.12.041 Reconstruction of the electrical sources of human EEG activity at high spatio-temporal accuracy is an important aim in neuroscience and neurological diagnostics. Over the last decades, numerous studies have demonstrated that realistic modeling of head anatomy improves the accuracy of source reconstruction of EEG signals. For example, including a cerebro-spinal fluid compartment and the anisotropy of white matter electrical conductivity were both shown to significantly reduce modeling errors. Here, we for the first time quantify the role of detailed reconstructions of the cerebral blood vessels in volume conductor head modeling for EEG. To study the role of the highly arborized cerebral blood vessels, we created a submillimeter head model based on ultra-high-field-strength (7T) structural MRI datasets. Blood vessels (arteries and emissary/intraosseous veins) were segmented using Frangi multi-scale vesselness filtering. The final head model consisted of a geometry-adapted cubic mesh with over 17×10(6) nodes. We solved the forward model using a finite-element-method (FEM) transfer matrix approach, which allowed reducing computation times substantially and quantified the importance of the blood vessel compartment by computing forward and inverse errors resulting from ignoring the blood vessels. Our results show that ignoring emissary veins piercing the skull leads to focal localization errors of approx. 5 to 15mm. Large errors (>2cm) were observed due to the carotid arteries and the dense arterial vasculature in areas such as in the insula or in the medial temporal lobe. Thus, in such predisposed areas, errors caused by neglecting blood vessels can reach similar magnitudes as those previously reported for neglecting white matter anisotropy, the CSF or the dura - structures which are generally considered important components of realistic EEG head models. Our findings thus imply that including a realistic blood vessel compartment in EEG head models will be helpful to improve the accuracy of EEG source analyses particularly when high accuracies in brain areas with dense vasculature are required. |
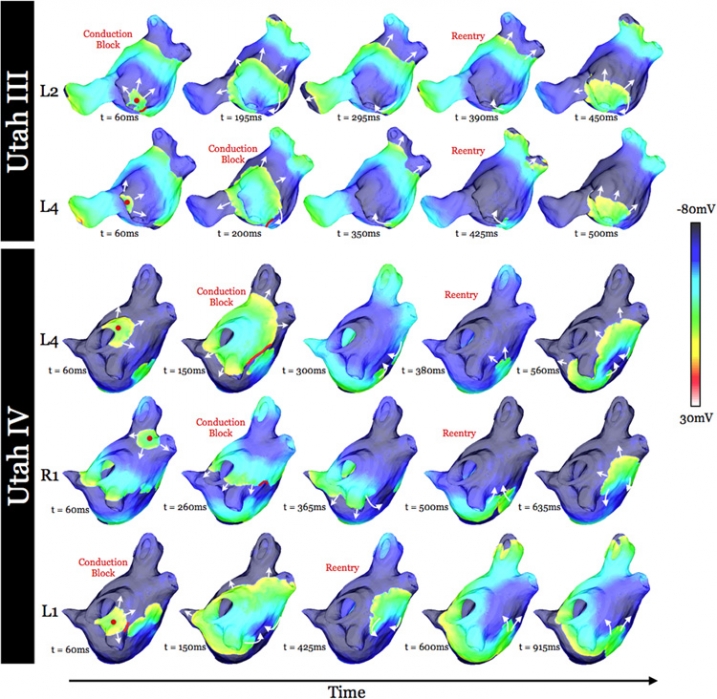
  Increased Susceptibility to Atrial Fibrillation Secondary to Atrial Fibrosis in Transgenic Goats Expressing Transforming Growth Factor-β1 I.A. Polejaeva, R. Ranjan, C.J. Davies, M. Regouski, J. Hall, A.L. Olsen, Q. Meng, H.M. Rutigliano, D.J. Dosdall, N.A. Angel, F.B. Sachse, T. Seidel, A.J. Thomas, R. Stott, K.E. Panter, P.M. Lee, A.J. Van Wettere, J.R. Stevens, Z. Wang, R.S. Macleod, N.F. Marrouche, K.L. White. In Journal of Cardiovascular Electrophysiology, Vol. 27, No. 10, Wiley-Blackwell, pp. 1220--1229. Aug, 2016. DOI: 10.1111/jce.13049 Introduction |
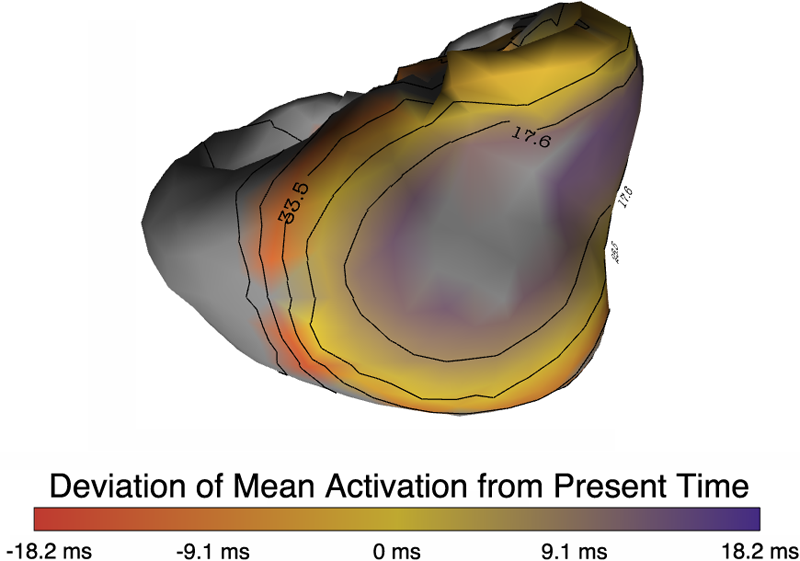
  muView: A Visual Analysis System for Exploring Uncertainty in Myocardial Ischemia Simulations P. Rosen, B. Burton, K. Potter, C.R. Johnson. In Visualization in Medicine and Life Sciences III, Springer Nature, pp. 49--69. 2016. DOI: 10.1007/978-3-319-24523-2_3 In this paper we describe the Myocardial Uncertainty Viewer (muView or µView) system for exploring data stemming from the simulation of cardiac ischemia. The simulation uses a collection of conductivity values to understand how ischemic regions effect the undamaged anisotropic heart tissue. The data resulting from the simulation is multi-valued and volumetric, and thus, for every data point, we have a collection of samples describing cardiac electrical properties. µView combines a suite of visual analysis methods to explore the area surrounding the ischemic zone and identify how perturbations of variables change the propagation of their effects. In addition to presenting a collection of visualization techniques, which individually highlight different aspects of the data, the coordinated view system forms a cohesive environment for exploring the simulations.We also discuss the findings of our study, which are helping to steer further development of the simulation and strengthening our collaboration with the biomedical engineers attempting to understand the phenomenon. |
  Virtual Electrophysiological Study of Atrial Fibrillation in Fibrotic Remodeling K. S. McDowell, S. Zahid, F. Vadakkumpadan, J. Blauer, R. S. MacLeod, N. A. Trayanova. In PLoS ONE, Vol. 10, No. 2, Public Library of Science, pp. 1-16. May, 2015. DOI: doi.org/10.1371/journal.pone.0117110 Research has indicated that atrial fibrillation (AF) ablation failure is related to the presence of atrial fibrosis. However it remains unclear whether this information can be successfully used in predicting the optimal ablation targets for AF termination. We aimed to provide a proof-of-concept that patient-specific virtual electrophysiological study that combines i) atrial structure and fibrosis distribution from clinical MRI and ii) modeling of atrial electrophysiology, could be used to predict: (1) how fibrosis distribution determines the locations from which paced beats degrade into AF; (2) the dynamic behavior of persistent AF rotors; and (3) the optimal ablation targets in each patient. Four MRI-based patient-specific models of fibrotic left atria were generated, ranging in fibrosis amount. Virtual electrophysiological studies were performed in these models, and where AF was inducible, the dynamics of AF were used to determine the ablation locations that render AF non-inducible. In 2 of the 4 models patient-specific models AF was induced; in these models the distance between a given pacing location and the closest fibrotic region determined whether AF was inducible from that particular location, with only the mid-range distances resulting in arrhythmia. Phase singularities of persistent rotors were found to move within restricted regions of tissue, which were independent of the pacing location from which AF was induced. Electrophysiological sensitivity analysis demonstrated that these regions changed little with variations in electrophysiological parameters. Patient-specific distribution of fibrosis was thus found to be a critical component of AF initiation and maintenance. When the restricted regions encompassing the meander of the persistent phase singularities were modeled as ablation lesions, AF could no longer be induced. The study demonstrates that a patient-specific modeling approach to identify non-invasively AF ablation targets prior to the clinical procedure is feasible. |
  The use of stimulation field models for deep brain stimulation programming C. R. Butson, C. C. McIntyre. In Brain Stimulation, Vol. 8, No. 5, Elsevier BV, pp. 976--978. September, 2015. DOI: 10.1016/j.brs.2015.06.005 |
  Proceedings of the Second Annual Deep Brain Stimulation Think Tank: What's in the Pipeline A. Gunduz, H. Morita, P. J. Rossi, W. L. Allen, R. L. Alterman, H. Bronte-Stewart, C. R. Butson, D. Charles, S. Deckers, C. de Hemptinne, M. DeLong, D. Dougherty, J. Ellrich, K. D. Foote, J. Giordano, W. Goodman, B. D. Greenberg, D. Greene, R. Gross, J. W. Judy, E. Karst, A. Kent, B. Kopell, A. Lang, A. Lozano, C. Lungu, K. E. Lyons, A. Machado, H. Martens, C. McIntyre, H. Min, J. Neimat, J. Ostrem, S. Pannu, F. Ponce, N. Pouratian, D. Reymers, L. Schrock, S. Sheth, L. Shih, S. Stanslaski, G. K. Steinke, P. Stypulkowski, A. I. Tröster, L. Verhagen, H. Walker, M. S. Okun. In International Journal of Neuroscience, Vol. 125, No. 7, Taylor & Francis, pp. 475-485. 2015. DOI: 10.3109/00207454.2014.999268 PubMed ID: 25526555 The proceedings of the 2nd Annual Deep Brain Stimulation Think Tank summarize the most contemporary clinical, electrophysiological, and computational work on DBS for the treatment of neurological and neuropsychiatric disease and represent the insights of a unique multidisciplinary ensemble of expert neurologists, neurosurgeons, neuropsychologists, psychiatrists, scientists, engineers and members of industry. Presentations and discussions covered a broad range of topics, including advocacy for DBS, improving clinical outcomes, innovations in computational models of DBS, understanding of the neurophysiology of Parkinson's disease (PD) and Tourette syndrome (TS) and evolving sensor and device technologies. |
 Poor scar formation after ablation is associated with atrial fibrillation recurrence, B.R. Parmar, T.R. Jarrett, E.G. Kholmovski, N. Hu, D. Parker, R.S. MacLeod, N.F. Marrouche, R. Ranjan. In Journal of Interventional Cardiac Electrophysiology, Vol. 44, No. 3, pp. 247-256. December, 2015. Purpose |
  A Kalman Filtering Perspective for Multiatlas Segmentation Y. Gao, L. Zhu, J. Cates, R. S. MacLeod, S. Bouix,, A. Tannenbaum. In SIAM J. Imaging Sciences, Vol. 8, No. 2, pp. 1007-1029. 2015. DOI: 10.1137/130933423 In multiatlas segmentation, one typically registers several atlases to the novel image, and their respective segmented label images are transformed and fused to form the final segmentation. In this work, we provide a new dynamical system perspective for multiatlas segmentation, inspired by the following fact: The transformation that aligns the current atlas to the novel image can be not only computed by direct registration but also inferred from the transformation that aligns the previous atlas to the image together with the transformation between the two atlases. This process is similar to the global positioning system on a vehicle, which gets position by inquiring from the satellite and by employing the previous location and velocity—neither answer in isolation being perfect. To solve this problem, a dynamical system scheme is crucial to combine the two pieces of information; for example, a Kalman filtering scheme is used. Accordingly, in this work, a Kalman multiatlas segmentation is proposed to stabilize the global/affine registration step. The contributions of this work are twofold. First, it provides a new dynamical systematic perspective for standard independent multiatlas registrations, and it is solved by Kalman filtering. Second, with very little extra computation, it can be combined with most existing multiatlas segmentation schemes for better registration/segmentation accuracy. |
  Virtual Electrophysiological Study of Atrial Fibrillation in Fibrotic Remodeling K.S. McDowell, S. Zahid, F. Vadakkumpadan, J.J. Blauer, R.S. MacLeod, N.A. Trayanova. In PLoS ONE, Vol. 10, No. 2, pp. e0117110. February, 2015. DOI: 10.1371/journal.pone.0117110 Research has indicated that atrial fibrillation (AF) ablation failure is related to the presence of atrial fibrosis. However it remains unclear whether this information can be successfully used in predicting the optimal ablation targets for AF termination. We aimed to provide a proof-of-concept that patient-specific virtual electrophysiological study that combines i) atrial structure and fibrosis distribution from clinical MRI and ii) modeling of atrial electrophysiology, could be used to predict: (1) how fibrosis distribution determines the locations from which paced beats degrade into AF; (2) the dynamic behavior of persistent AF rotors; and (3) the optimal ablation targets in each patient. Four MRI-based patient-specific models of fibrotic left atria were generated, ranging in fibrosis amount. Virtual electrophysiological studies were performed in these models, and where AF was inducible, the dynamics of AF were used to determine the ablation locations that render AF non-inducible. In 2 of the 4 models patient-specific models AF was induced; in these models the distance between a given pacing location and the closest fibrotic region determined whether AF was inducible from that particular location, with only the mid-range distances resulting in arrhythmia. Phase singularities of persistent rotors were found to move within restricted regions of tissue, which were independent of the pacing location from which AF was induced. Electrophysiological sensitivity analysis demonstrated that these regions changed little with variations in electrophysiological parameters. Patient-specific distribution of fibrosis was thus found to be a critical component of AF initiation and maintenance. When the restricted regions encompassing the meander of the persistent phase singularities were modeled as ablation lesions, AF could no longer be induced. The study demonstrates that a patient-specific modeling approach to identify non-invasively AF ablation targets prior to the clinical procedure is feasible. |
 Scientific Visualization: Uncertainty, Multifield, Biomedical, and Scalable Visualization, C.D. Hansen, M. Chen, C.R. Johnson, A.E. Kaufman, H. Hagen (Eds.). Mathematics and Visualization, Springer, 2014. ISBN: 978-1-4471-6496-8 |
  SVD Identifies Transcript Length Distribution Functions from DNA Microarray Data and Reveals Evolutionary Forces Globally Affecting GBM Metabolism N.M. Bertagnolli, J.A. Drake, J.M. Tennessen, O. Alter. In Public Library of Science (PLoS) One, Vol. 8, No. 11, pp. article e78913. November, 2013. DOI: 10.1371/journal.pone.0078913 To search for evolutionary forces that might act upon transcript length, we use the singular value decomposition (SVD) to identify the length distribution functions of sets and subsets of human and yeast transcripts from profiles of mRNA abundance levels across gel electrophoresis migration distances that were previously measured by DNA microarrays. We show that the SVD identifies the transcript length distribution functions as “asymmetric generalized coherent states” from the DNA microarray data and with no a-priori assumptions. Comparing subsets of human and yeast transcripts of the same gene ontology annotations, we find that in both disparate eukaryotes, transcripts involved in protein synthesis or mitochondrial metabolism are significantly shorter than typical, and in particular, significantly shorter than those involved in glucose metabolism. Comparing the subsets of human transcripts that are overexpressed in glioblastoma multiforme (GBM) or normal brain tissue samples from The Cancer Genome Atlas, we find that GBM maintains normal brain overexpression of significantly short transcripts, enriched in transcripts that are involved in protein synthesis or mitochondrial metabolism, but suppresses normal overexpression of significantly longer transcripts, enriched in transcripts that are involved in glucose metabolism and brain activity. These global relations among transcript length, cellular metabolism and tumor development suggest a previously unrecognized physical mode for tumor and normal cells to differentially regulate metabolism in a transcript length-dependent manner. The identified distribution functions support a previous hypothesis from mathematical modeling of evolutionary forces that act upon transcript length in the manner of the restoring force of the harmonic oscillator. |
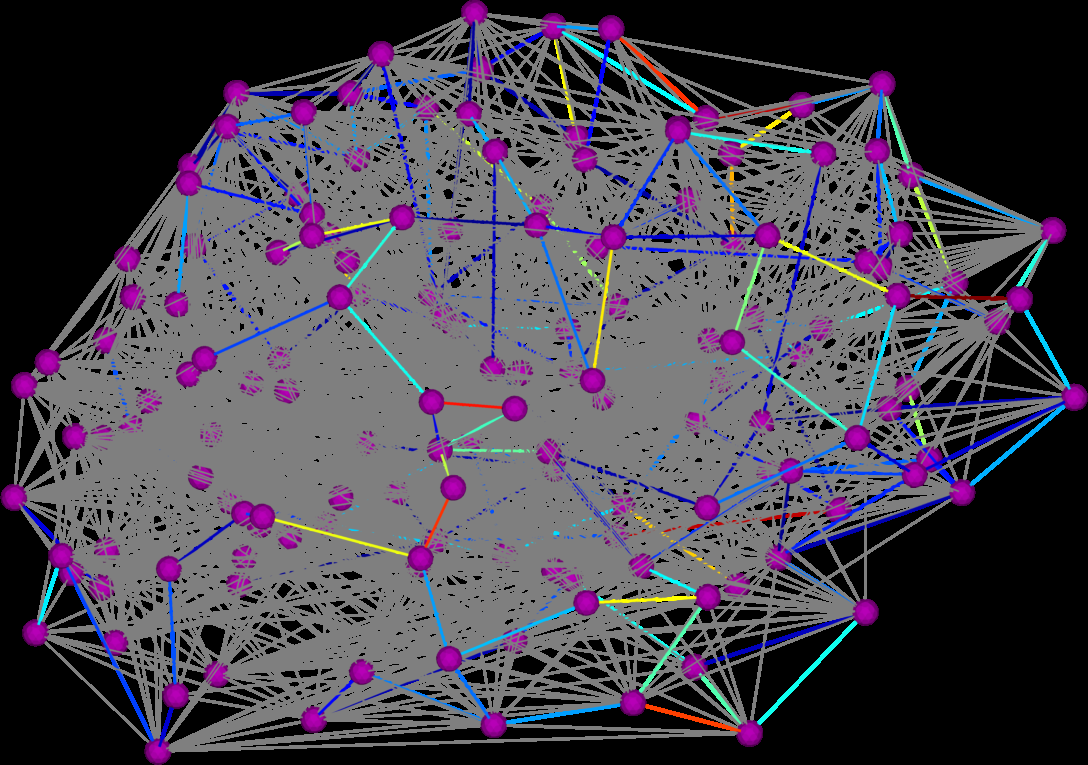
  Graph Diffusion Distance: A Difference Measure for Weighted Graphs Based on the Graph Laplacian Exponential Kernel D.K. Hammond, Y. Gur, C.R. Johnson. In Proceedings of the IEEE global conference on information and signal processing (GlobalSIP'13), Austin, Texas, pp. 419--422. 2013. DOI: 10.1109/GlobalSIP.2013.6736904 We propose a novel difference metric, called the graph diffusion distance (GDD), for quantifying the difference between two weighted graphs with the same number of vertices. Our approach is based on measuring the average similarity of heat diffusion on each graph. We compute the graph Laplacian exponential kernel matrices, corresponding to repeatedly solving the heat diffusion problem with initial conditions localized to single vertices. The GDD is then given by the Frobenius norm of the difference of the kernels, at the diffusion time yielding the maximum difference. We study properties of the proposed distance on both synthetic examples, and on real-data graphs representing human anatomical brain connectivity. |
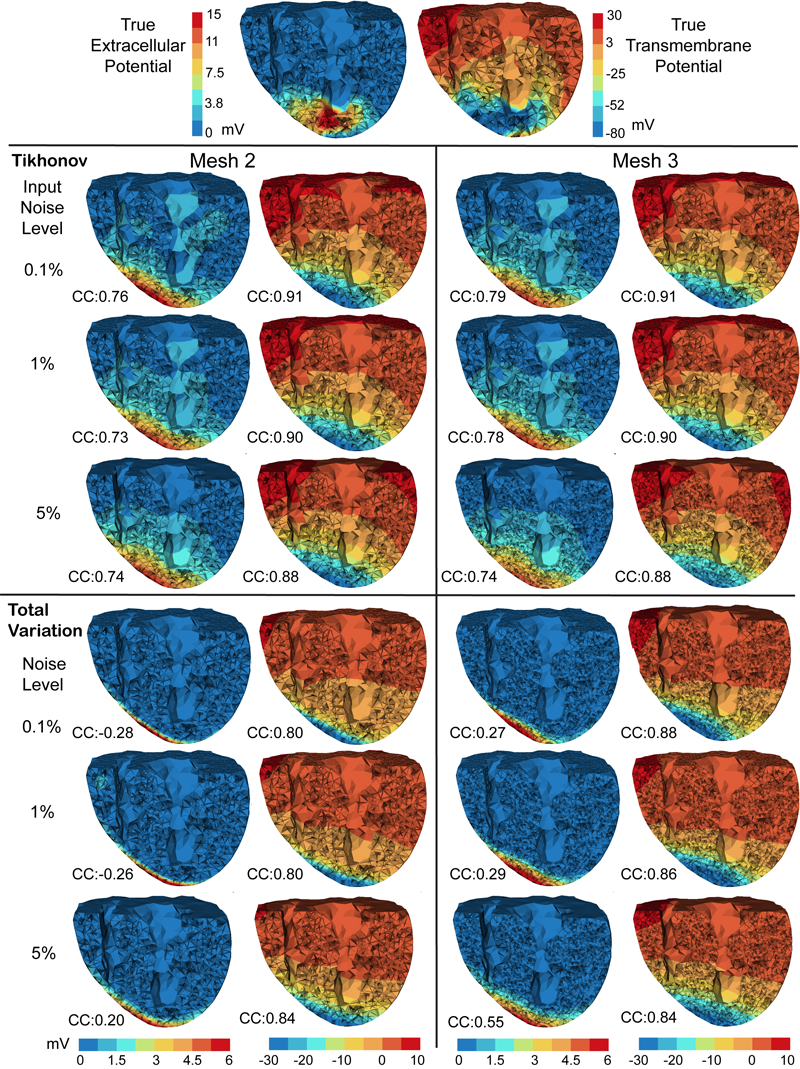
  Inverse Electrocardiographic Source Localization of Ischemia: An Optimization Framework and Finite Element Solution D. Wang, R.M. Kirby, R.S. MacLeod, C.R. Johnson. In Journal of Computational Physics, Vol. 250, Academic Press, pp. 403--424. 2013. ISSN: 0021-9991 DOI: 10.1016/j.jcp.2013.05.027 With the goal of non-invasively localizing cardiac ischemic disease using bodysurface potential recordings, we attempted to reconstruct the transmembrane potential (TMP) throughout the myocardium with the bidomain heart model. The task is an inverse source problem governed by partial differential equations (PDE). Our main contribution is solving the inverse problem within a PDE-constrained optimization framework that enables various physically-based constraints in both equality and inequality forms. We formulated the optimality conditions rigorously in the continuum before deriving finite element discretization, thereby making the optimization independent of discretization choice. Such a formulation was derived for the L2-norm Tikhonov regularization and the total variation minimization. The subsequent numerical optimization was fulfilled by a primal-dual interior-point method tailored to our problem's specific structure. Our simulations used realistic, fiberincluded heart models consisting of up to 18,000 nodes, much finer than any inverse models previously reported. With synthetic ischemia data we localized ischemic regions with roughly a 10% false-negative rate or a 20% false-positive rate under conditions up to 5% input noise. With ischemia data measured from animal experiments, we reconstructed TMPs with roughly 0.9 correlation with the ground truth. While precisely estimating the TMP in general cases remains an open problem, our study shows the feasibility of reconstructing TMP during the ST interval as a means of ischemia localization. Keywords: cvrti, 2P41 GM103545-14 |