Image Analysis
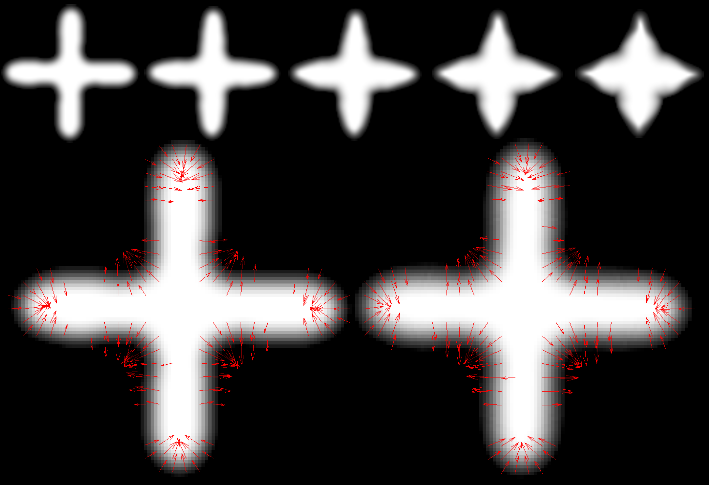
SCI's imaging work addresses fundamental questions in 2D and 3D image processing, including filtering, segmentation, surface reconstruction, and shape analysis. In low-level image processing, this effort has produce new nonparametric methods for modeling image statistics, which have resulted in better algorithms for denoising and reconstruction. Work with particle systems has led to new methods for visualizing and analyzing 3D surfaces. Our work in image processing also includes applications of advanced computing to 3D images, which has resulted in new parallel algorithms and real-time implementations on graphics processing units (GPUs). Application areas include medical image analysis, biological image processing, defense, environmental monitoring, and oil and gas.
Ross Whitaker
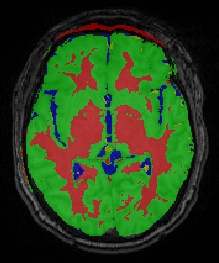
Segmentation
Chris Johnson
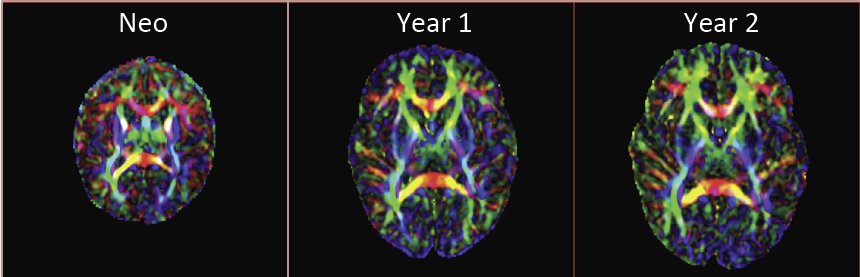
Diffusion Tensor AnalysisFunded Research Projects:
Publications in Image Analysis:
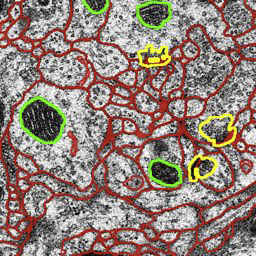
  Multi-class Multi-scale Series Contextual Model for Image Segmentation M. Seyedhosseini, T. Tasdizen. In IEEE Transactions on Image Processing, Vol. PP, No. 99, 2013. DOI: 10.1109/TIP.2013.2274388 Contextual information has been widely used as a rich source of information to segment multiple objects in an image. A contextual model utilizes the relationships between the objects in a scene to facilitate object detection and segmentation. However, using contextual information from different objects in an effective way for object segmentation remains a difficult problem. In this paper, we introduce a novel framework, called multi-class multi-scale (MCMS) series contextual model, which uses contextual information from multiple objects and at different scales for learning discriminative models in a supervised setting. The MCMS model incorporates cross-object and inter-object information into one probabilistic framework and thus is able to capture geometrical relationships and dependencies among multiple objects in addition to local information from each single object present in an image. We demonstrate that our MCMS model improves object segmentation performance in electron microscopy images and provides a coherent segmentation of multiple objects. By speeding up the segmentation process, the proposed method will allow neurobiologists to move beyond individual specimens and analyze populations paving the way for understanding neurodegenerative diseases at the microscopic level. |
  Modeling 4D changes in pathological anatomy using domain adaptation: analysis of TBI imaging using a tumor database Bo Wang, M. Prastawa, A. Saha, S.P. Awate, A. Irimia, M.C. Chambers, P.M. Vespa, J.D. Van Horn, V. Pascucci, G. Gerig. In Proceedings of the 2013 MICCAI-MBIA Workshop, Lecture Notes in Computer Science (LNCS), Vol. 8159, Note: Awarded Best Paper!, pp. 31--39. 2013. DOI: 10.1007/978-3-319-02126-3_4 Analysis of 4D medical images presenting pathology (i.e., lesions) is signi cantly challenging due to the presence of complex changes over time. Image analysis methods for 4D images with lesions need to account for changes in brain structures due to deformation, as well as the formation and deletion of new structures (e.g., edema, bleeding) due to the physiological processes associated with damage, intervention, and recovery. We propose a novel framework that models 4D changes in pathological anatomy across time, and provides explicit mapping from a healthy template to subjects with pathology. Moreover, our framework uses transfer learning to leverage rich information from a known source domain, where we have a collection of completely segmented images, to yield effective appearance models for the input target domain. The automatic 4D segmentation method uses a novel domain adaptation technique for generative kernel density models to transfer information between different domains, resulting in a fully automatic method that requires no user interaction. We demonstrate the effectiveness of our novel approach with the analysis of 4D images of traumatic brain injury (TBI), using a synthetic tumor database as the source domain. |
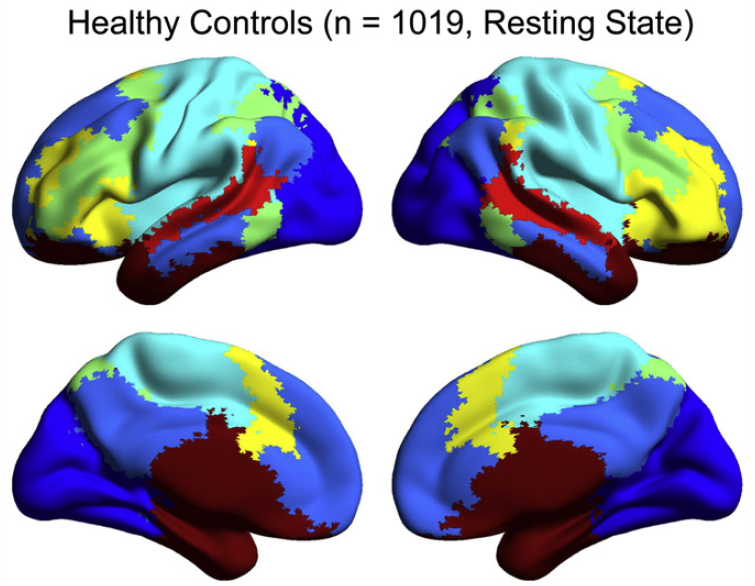
  Abnormal brain synchrony in Down Syndrome, J.S. Anderson, J.A. Nielsen, M.A. Ferguson, M.C. Burback, E.T. Cox, L. Dai, G. Gerig, J.O. Edgin, J.R. Korenberg. In NeuroImage: Clinical, Vol. 2, pp. 703--715. 2013. ISSN: 2213-1582 DOI: 10.1016/j.nicl.2013.05.006 Down Syndrome is the most common genetic cause for intellectual disability, yet the pathophysiology of cognitive impairment in Down Syndrome is unknown. We compared fMRI scans of 15 individuals with Down Syndrome to 14 typically developing control subjects while they viewed 50 min of cartoon video clips. There was widespread increased synchrony between brain regions, with only a small subset of strong, distant connections showing underconnectivity in Down Syndrome. Brain regions showing negative correlations were less anticorrelated and were among the most strongly affected connections in the brain. Increased correlation was observed between all of the distributed brain networks studied, with the strongest internetwork correlation in subjects with the lowest performance IQ. A functional parcellation of the brain showed simplified network structure in Down Syndrome organized by local connectivity. Despite increased interregional synchrony, intersubject correlation to the cartoon stimuli was lower in Down Syndrome, indicating that increased synchrony had a temporal pattern that was not in response to environmental stimuli, but idiosyncratic to each Down Syndrome subject. Short-range, increased synchrony was not observed in a comparison sample of 447 autism vs. 517 control subjects from the Autism Brain Imaging Exchange (ABIDE) collection of resting state fMRI data, and increased internetwork synchrony was only observed between the default mode and attentional networks in autism. These findings suggest immature development of connectivity in Down Syndrome with impaired ability to integrate information from distant brain regions into coherent distributed networks. |
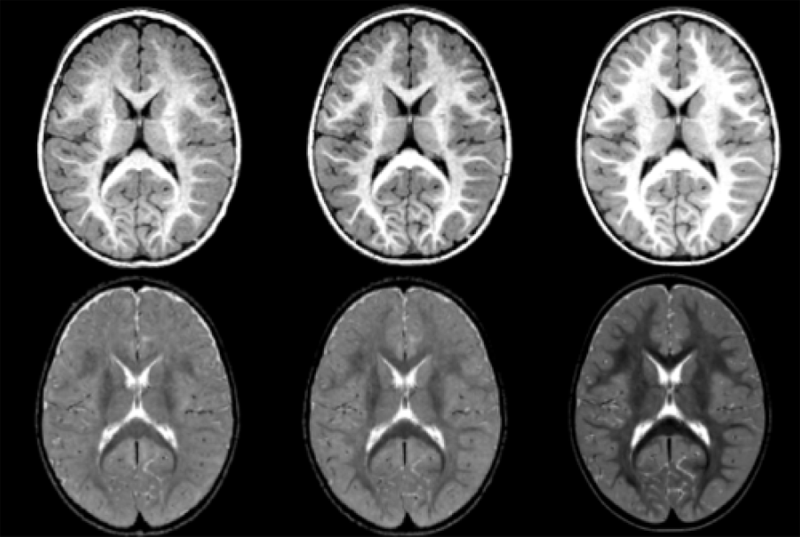
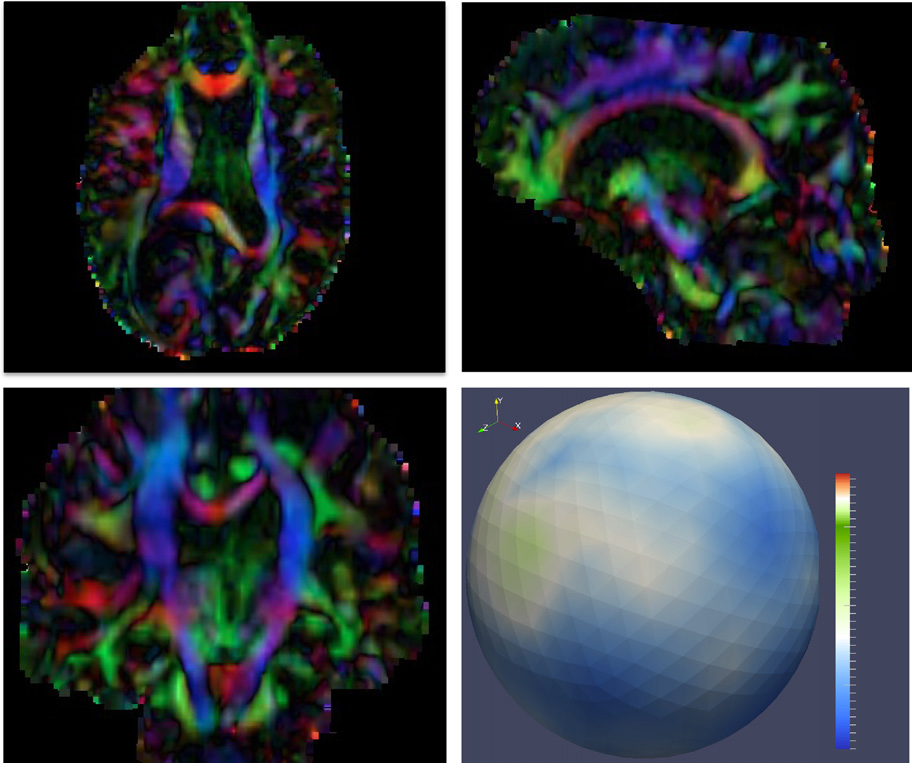
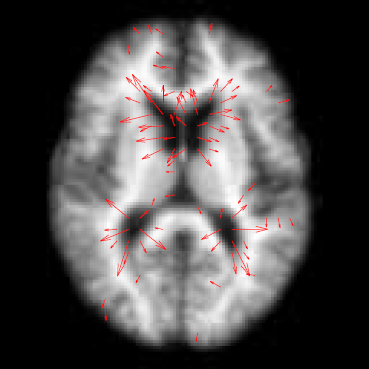
  Diffusion imaging quality control via entropy of principal direction distribution, M. Farzinfar, I. Oguz, R.G. Smith, A.R. Verde, C. Dietrich, A. Gupta, M.L. Escolar, J. Piven, S. Pujol, C. Vachet, S. Gouttard, G. Gerig, S. Dager, R.C. McKinstry, S. Paterson, A.C. Evans, M.A. Styner. In NeuroImage, Vol. 82, pp. 1--12. 2013. ISSN: 1053-8119 DOI: 10.1016/j.neuroimage.2013.05.022 Diffusion MR imaging has received increasing attention in the neuroimaging community, as it yields new insights into the microstructural organization of white matter that are not available with conventional MRI techniques. While the technology has enormous potential, diffusion MRI suffers from a unique and complex set of image quality problems, limiting the sensitivity of studies and reducing the accuracy of findings. Furthermore, the acquisition time for diffusion MRI is longer than conventional MRI due to the need for multiple acquisitions to obtain directionally encoded Diffusion Weighted Images (DWI). This leads to increased motion artifacts, reduced signal-to-noise ratio (SNR), and increased proneness to a wide variety of artifacts, including eddy-current and motion artifacts, “venetian blind” artifacts, as well as slice-wise and gradient-wise inconsistencies. Such artifacts mandate stringent Quality Control (QC) schemes in the processing of diffusion MRI data. Most existing QC procedures are conducted in the DWI domain and/or on a voxel level, but our own experiments show that these methods often do not fully detect and eliminate certain types of artifacts, often only visible when investigating groups of DWI's or a derived diffusion model, such as the most-employed diffusion tensor imaging (DTI). Here, we propose a novel regional QC measure in the DTI domain that employs the entropy of the regional distribution of the principal directions (PD). The PD entropy quantifies the scattering and spread of the principal diffusion directions and is invariant to the patient's position in the scanner. High entropy value indicates that the PDs are distributed relatively uniformly, while low entropy value indicates the presence of clusters in the PD distribution. The novel QC measure is intended to complement the existing set of QC procedures by detecting and correcting residual artifacts. Such residual artifacts cause directional bias in the measured PD and here called dominant direction artifacts. Experiments show that our automatic method can reliably detect and potentially correct such artifacts, especially the ones caused by the vibrations of the scanner table during the scan. The results further indicate the usefulness of this method for general quality assessment in DTI studies. Keywords: Diffusion magnetic resonance imaging, Diffusion tensor imaging, Quality assessment, Entropy |
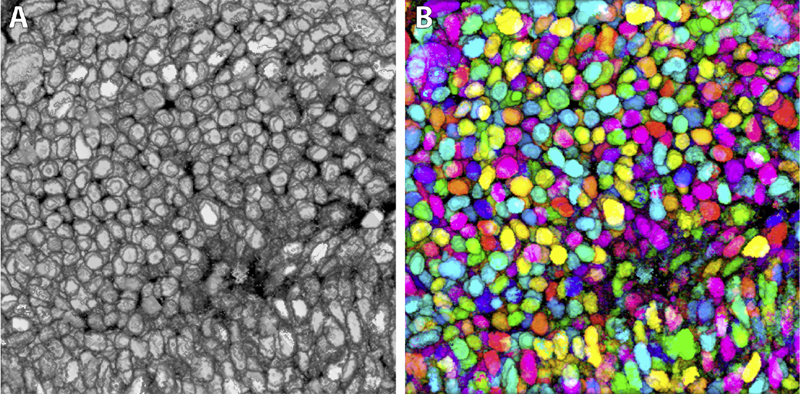
  Synthetic Brainbows Y. Wan, H. Otsuna, C.D. Hansen. In Computer Graphics Forum, Vol. 32, No. 3pt4, Wiley-Blackwell, pp. 471--480. jun, 2013. DOI: 10.1111/cgf.12134 Brainbow is a genetic engineering technique that randomly colorizes cells. Biological samples processed with this technique and imaged with confocal microscopy have distinctive colors for individual cells. Complex cellular structures can then be easily visualized. However, the complexity of the Brainbow technique limits its applications. In practice, most confocal microscopy scans use different florescence staining with typically at most three distinct cellular structures. These structures are often packed and obscure each other in rendered images making analysis difficult. In this paper, we leverage a process known as GPU framebuffer feedback loops to synthesize Brainbow-like images. In addition, we incorporate ID shuffling and Monte-Carlo sampling into our technique, so that it can be applied to single-channel confocal microscopy data. The synthesized Brainbow images are presented to domain experts with positive feedback. A user survey demonstrates that our synthetic Brainbow technique improves visualizations of volume data with complex structures for biologists. |
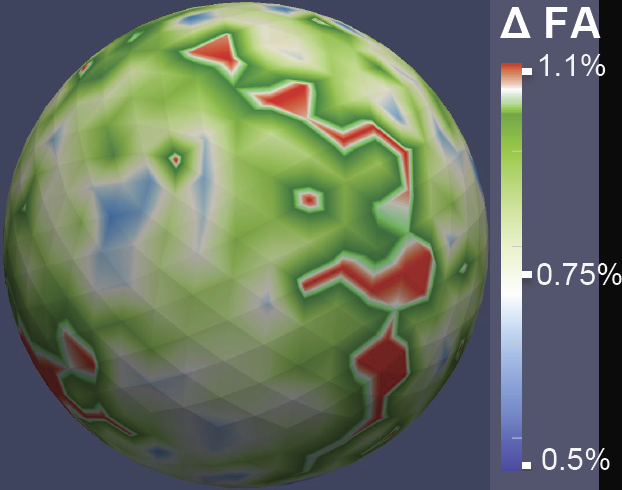
  DTI Quality Control Assessment via Error Estimation From Monte Carlo Simulations M. Farzinfar, Y. Li, A.R. Verde, I. Oguz, G. Gerig, M.A. Styner. In Proceedings of SPIE 8669, Medical Imaging 2013: Image Processing, Vol. 8669, 2013. DOI: 10.1117/12.2006925 PubMed ID: 23833547 PubMed Central ID: PMC3702180 Diffusion Tensor Imaging (DTI) is currently the state of the art method for characterizing the microscopic tissue structure of white matter in normal or diseased brain in vivo. DTI is estimated from a series of Diffusion Weighted Imaging (DWI) volumes. DWIs suffer from a number of artifacts which mandate stringent Quality Control (QC) schemes to eliminate lower quality images for optimal tensor estimation. Conventionally, QC procedures exclude artifact-affected DWIs from subsequent computations leading to a cleaned, reduced set of DWIs, called DWI-QC. Often, a rejection threshold is heuristically/empirically chosen above which the entire DWI-QC data is rendered unacceptable and thus no DTI is computed. In this work, we have devised a more sophisticated, Monte-Carlo (MC) simulation based method for the assessment of resulting tensor properties. This allows for a consistent, error-based threshold definition in order to reject/accept the DWI-QC data. Specifically, we propose the estimation of two error metrics related to directional distribution bias of Fractional Anisotropy (FA) and the Principal Direction (PD). The bias is modeled from the DWI-QC gradient information and a Rician noise model incorporating the loss of signal due to the DWI exclusions. Our simulations further show that the estimated bias can be substantially different with respect to magnitude and directional distribution depending on the degree of spatial clustering of the excluded DWIs. Thus, determination of diffusion properties with minimal error requires an evenly distributed sampling of the gradient directions before and after QC. |
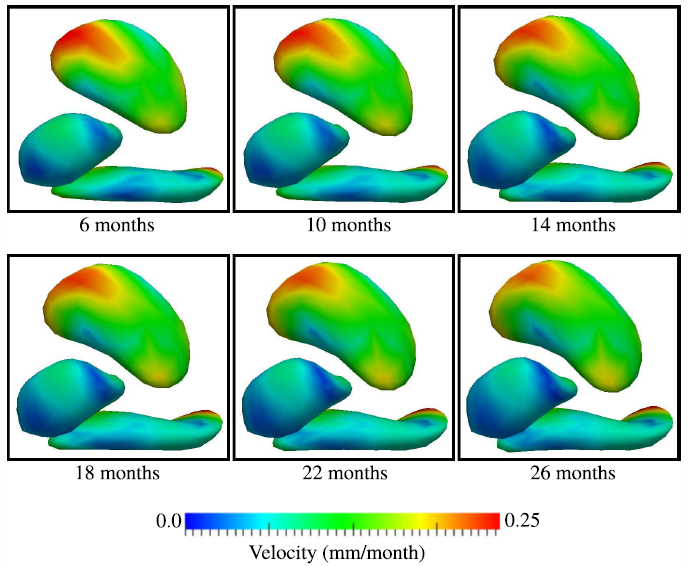
  3D of brain shape and volume after cranial vault remodeling surgery for craniosynostosis correction in infants B. Paniagua, O. Emodi, J. Hill, J. Fishbaugh, L.A. Pimenta, S.R. Aylward, E. Andinet, G. Gerig, J. Gilmore, J.A. van Aalst, M. Styner. In Proceedings of SPIE 8672, Medical Imaging 2013: Biomedical Applications in Molecular, Structural, and Functional Imaging, 86720V, 2013. DOI: 10.1117/12.2006524 The skull of young children is made up of bony plates that enable growth. Craniosynostosis is a birth defect that causes one or more sutures on an infant’s skull to close prematurely. Corrective surgery focuses on cranial and orbital rim shaping to return the skull to a more normal shape. Functional problems caused by craniosynostosis such as speech and motor delay can improve after surgical correction, but a post-surgical analysis of brain development in comparison with age-matched healthy controls is necessary to assess surgical outcome. Full brain segmentations obtained from pre- and post-operative computed tomography (CT) scans of 8 patients with single suture sagittal (n=5) and metopic (n=3), nonsyndromic craniosynostosis from 41 to 452 days-of-age were included in this study. Age-matched controls obtained via 4D acceleration-based regression of a cohort of 402 full brain segmentations from healthy controls magnetic resonance images (MRI) were also used for comparison (ages 38 to 825 days). 3D point-based models of patient and control cohorts were obtained using SPHARM-PDM shape analysis tool. From a full dataset of regressed shapes, 240 healthy regressed shapes between 30 and 588 days-of-age (time step = 2.34 days) were selected. Volumes and shape metrics were obtained for craniosynostosis and healthy age-matched subjects. Volumes and shape metrics in single suture craniosynostosis patients were larger than age-matched controls for pre- and post-surgery. The use of 3D shape and volumetric measurements show that brain growth is not normal in patients with single suture craniosynostosis. |
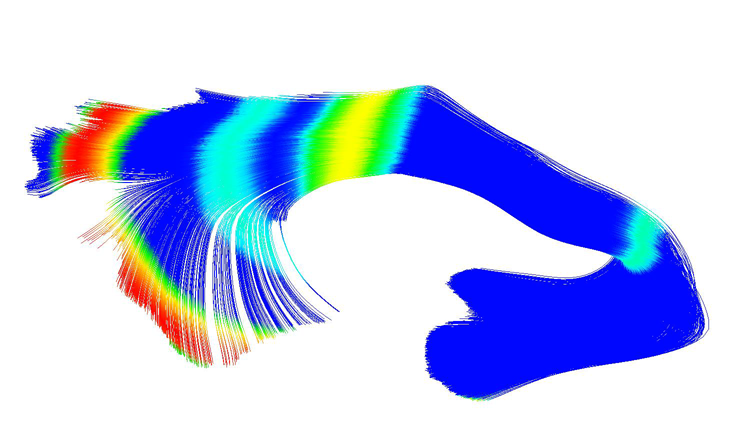
  UNC-Utah NA-MIC DTI framework: atlas based fiber tract analysis with application to a study of nicotine smoking addiction A.R. Verde, J.-B. Berger, A. Gupta, M. Farzinfar, A. Kaiser, V.W. Chanon, C. Boettiger, H. Johnson, J. Matsui, A. Sharma, C. Goodlett, Y. Shi, H. Zhu, G. Gerig, S. Gouttard, C. Vachet, M. Styner. In Proc. SPIE 8669, Medical Imaging 2013: Image Processing, 86692D, Vol. 8669, pp. 86692D-86692D-8. 2013. DOI: 10.1117/12.2007093 Purpose: The UNC-Utah NA-MIC DTI framework represents a coherent, open source, atlas fiber tract based DTI analysis framework that addresses the lack of a standardized fiber tract based DTI analysis workflow in the field. Most steps utilize graphical user interfaces (GUI) to simplify interaction and provide an extensive DTI analysis framework for non-technical researchers/investigators. Data: We illustrate the use of our framework on a 54 directional DWI neuroimaging study contrasting 15 Smokers and 14 Controls. Method(s): At the heart of the framework is a set of tools anchored around the multi-purpose image analysis platform 3D-Slicer. Several workflow steps are handled via external modules called from Slicer in order to provide an integrated approach. Our workflow starts with conversion from DICOM, followed by thorough automatic and interactive quality control (QC), which is a must for a good DTI study. Our framework is centered around a DTI atlas that is either provided as a template or computed directly as an unbiased average atlas from the study data via deformable atlas building. Fiber tracts are defined via interactive tractography and clustering on that atlas. DTI fiber profiles are extracted automatically using the atlas mapping information. These tract parameter profiles are then analyzed using our statistics toolbox (FADTTS). The statistical results are then mapped back on to the fiber bundles and visualized with 3D Slicer. Results: This framework provides a coherent set of tools for DTI quality control and analysis. Conclusions: This framework will provide the field with a uniform process for DTI quality control and analysis. |
  Model Selection and Estimation of Multi-Compartment Models in Diffusion MRI with a Rician Noise Model X. Zhu, Y. Gur, W. Wang, P.T. Fletcher. In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI), Lecture Notes in Computer Science (LNCS), Vol. 23, pp. 644--655. 2013. PubMed ID: 24684006 Multi-compartment models in diffusion MRI (dMRI) are used to describe complex white matter fiber architecture of the brain. In this paper, we propose a novel multi-compartment estimation method based on the ball-and-stick model, which is composed of an isotropic diffusion compartment (\"ball\") as well as one or more perfectly linear diffusion compartments (\"sticks\"). To model the noise distribution intrinsic to dMRI measurements, we introduce a Rician likelihood term and estimate the model parameters by means of an Expectation Maximization (EM) algorithm. This paper also addresses the problem of selecting the number of fiber compartments that best fit the data, by introducing a sparsity prior on the volume mixing fractions. This term provides automatic model selection and enables us to discriminate different fiber populations. When applied to simulated data, our method provides accurate estimates of the fiber orientations, diffusivities, and number of compartments, even at low SNR, and outperforms similar methods that rely on a Gaussian noise distribution assumption. We also apply our method to in vivo brain data and show that it can successfully capture complex fiber structures that match the known anatomy. |
  Geodesic Shape Regression in the Framework of Currents J. Fishbaugh, M.W. Prastawa, G. Gerig, S. Durrleman. In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI), Vol. 23, pp. 718--729. 2013. PubMed ID: 24684012 PubMed Central ID: PMC4127488 Shape regression is emerging as an important tool for the statistical analysis of time dependent shapes. In this paper, we develop a new generative model which describes shape change over time, by extending simple linear regression to the space of shapes represented as currents in the large deformation diffeomorphic metric mapping (LDDMM) framework. By analogy with linear regression, we estimate a baseline shape (intercept) and initial momenta (slope) which fully parameterize the geodesic shape evolution. This is in contrast to previous shape regression methods which assume the baseline shape is fixed. We further leverage a control point formulation, which provides a discrete and low dimensional parameterization of large diffeomorphic transformations. This flexible system decouples the parameterization of deformations from the specific shape representation, allowing the user to define the dimension- ality of the deformation parameters. We present an optimization scheme that estimates the baseline shape, location of the control points, and initial momenta simultaneously via a single gradient descent algorithm. Finally, we demonstrate our proposed method on synthetic data as well as real anatomical shape complexes. |
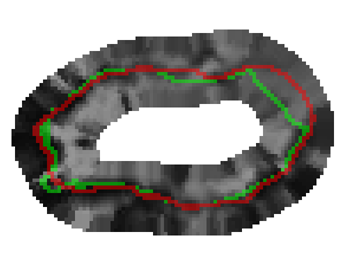
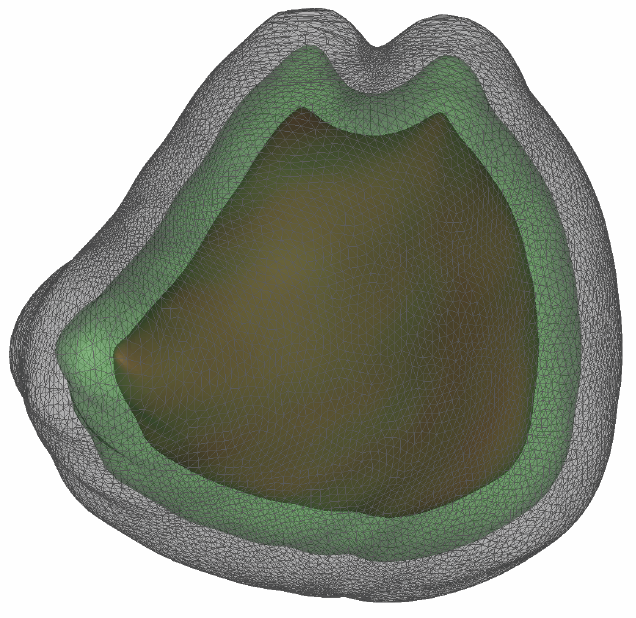
  Bayesian Segmentation of Atrium Wall using Globally-Optimal Graph Cuts on 3D Meshes G. Veni, S. Awate, Z. Fu, R.T. Whittaker. In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI), Lecture Notes in Computer Science (LNCS), Vol. 23, pp. 656--677. 2013. PubMed ID: 24684007 Efficient segmentation of the left atrium (LA) wall from delayed enhancement MRI is challenging due to inconsistent contrast, combined with noise, and high variation in atrial shape and size. We present a surface-detection method that is capable of extracting the atrial wall by computing an optimal a-posteriori estimate. This estimation is done on a set of nested meshes, constructed from an ensemble of segmented training images, and graph cuts on an associated multi-column, proper-ordered graph. The graph/mesh is a part of a template/model that has an associated set of learned intensity features. When this mesh is overlaid onto a test image, it produces a set of costs which lead to an optimal segmentation. The 3D mesh has an associated weighted, directed multi-column graph with edges that encode smoothness and inter-surface penalties. Unlike previous graph-cut methods that impose hard constraints on the surface properties, the proposed method follows from a Bayesian formulation resulting in soft penalties on spatial variation of the cuts through the mesh. The novelty of this method also lies in the construction of proper-ordered graphs on complex shapes for choosing among distinct classes of base shapes for automatic LA segmentation. We evaluate the proposed segmentation framework on simulated and clinical cardiac MRI. Keywords: Atrial Fibrillation, Bayesian segmentation, Minimum s-t cut, Mesh Generation, Geometric Graph |
  A Vector Momenta Formulation of Diffeomorphisms for Improved Geodesic Regression and Atlas Construction N.P. Singh, J. Hinkle, S. Joshi, P.T. Fletcher. In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), Note: Received Best Student Paper Award, pp. 1219--1222. 2013. DOI: 10.1109/ISBI.2013.6556700 This paper presents a novel approach for diffeomorphic image regression and atlas estimation that results in improved convergence and numerical stability. We use a vector momenta representation of a diffeomorphism's initial conditions instead of the standard scalar momentum that is typically used. The corresponding variational problem results in a closed form update for template estimation in both the geodesic regression and atlas estimation problems. While we show that the theoretical optimal solution is equivalent to the scalar momenta case, the simplification of the optimization problem leads to more stable and efficient estimation in practice. We demonstrate the effectiveness of our method for atlas estimation and geodesic regression using synthetically generated shapes and 3D MRI brain scans. Keywords: LDDMM, Geodesic regression, Atlas, Vector Momentum |
  Proper Ordered Meshing of Complex Shapes and Optimal Graph Cuts Applied to Atrial-Wall Segmentation from DE-MRI G. Veni, Z. Fu, S.P. Awate, R.T. Whitaker. In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 1296--1299. 2013. DOI: 10.1109/ISBI.2013.6556769 Segmentation of the left atrium wall from delayed enhancement MRI is challenging because of inconsistent contrast combined with noise and high variation in atrial shape and size. This paper presents a method for left-atrium wall segmentation by using a novel sophisticated mesh-generation strategy and graph cuts on a proper ordered graph. The mesh is part of a template/model that has an associated set of learned intensity features. When this mesh is overlaid onto a test image, it produces a set of costs on the graph vertices which eventually leads to an optimal segmentation. The novelty also lies in the construction of proper ordered graphs on complex shapes and for choosing among distinct classes of base shapes/meshes for automatic segmentation. We evaluate the proposed segmentation framework quantitatively on simulated and clinical cardiac MRI. |