The traditional method of validating a simulation is to identify a problem
that is simple enough that an analytical or closed form solution exists.
Then one can compare a numerically computed solution with the analytically
known true result. At a minimum, comparison of results from the analytical
and numerical solutions can be useful for identifying fundamental problems
with either the concept or the implementation of the more general numerical
approach. Because of their simplicity, however, analytical solutions may
provide data that establish only a useful set of validation
criteria, but clearly not a sufficient set. Any tenable numerical
approach must mimic the analytical result, but this is not enough to ensure
accuracy of the numerical approach under other, more realistic conditions.
Computational cross validation may also be possible in cases in which two
separate numerical or computational approaches can solve the same physical
problem. Here, too, the range of application of one of the computational
approaches may be limited so that validation is incomplete. For example,
one can implement a discrete source simulation to generate potentials on both
heart and torso surfaces and use the results to cross validate a different
inverse solution that computes heart potentials from torso potentials for
any source. The surface-to-surface solution is then validated only
for the dipole source and not for the broad range of applications for which
it was conceived. We provide more examples of both these approaches in
this section.
One significant advantage of most analytical or computational validation
approaches in inverse problems is the relative ease with which one can
control the essential factors such as geometric accuracy or level of detail
and--sometimes to a more limited extent--the source configuration. In
general, on the other hand, the geometry must remain simple in order for
the analytic solution to exist. Even for simple shapes, it is sometimes
possible to vary aspects of the geometry to at least begin to approach some
aspects of realistic conditions and thus permit some general statements
about the associated cardiac conditions. For example, one can use sets of
nested spheres to represent external and internal boundaries in the
volume conductor and then vary the conductivity assigned to each
region. Rudy et al. have shown that one can also impose some eccentricity
to the position of the spheres and thus suppress some of the complete
symmetry that limits concentric sphere models.[5,6]
The ease with which one can handle the second, more difficult requirement
for validation-the availability of source values and remote measured
potentials-is the main attraction of these methods; however, fidelity to
biological conditions is often quite limited.
The best known analytically tractable models in electrocardiology are the
concentric and eccentric spheres models proposed by Bayley and
Berry,[7,8,9] developed extensively by Rudy
et al.,[6,10,11] and still in use
today.[3,12,13,14,15,16]
Rudy et al. used these models for more than simply validating their
numerical solution. By varying source types and locations and geometric
model eccentricity, error, and conductivity, as well as regularization
functionals used for the inverse solutions, they developed several more
fundamental hypotheses about the relationships between each of these
factors and inverse solution accuracy. Some of these ideas have
subsequently been validated by physical models and human clinical studies.
However, the basic limitation of analytical models based on simple
geometries remains the uncertainty about the influence of real anatomical
structures. This, in turn, restricts the application of conclusions drawn
from such studies to realistic physiologic situations.
One validation approach that permits more realism in the geometric model is
to begin with simulated or measured source data, and then place them in a
realistic, discrete geometric model. These sources and a numerical forward
solution then provide synthetic torso data to which one can (must) add
noise, and then apply inverse solutions. Validation then consists of
comparing the inverse solutions to the known sources. The simulated source
data can, for example, be calculated from dipole
models,[17,18] taken from the
literature,[19]
measured in open chest or torso tank experiments[20,21,22,23,24,25] (see next
section), or calculated by an initial inverse solution from measured torso
surface data.[26,27,28,29,30]
The justification for this rather suspiciously circular validation strategy
comes from the fact that electrocardiographic inverse solutions are much
less accurate than the associated forward solutions. The main challenge of
the inverse problem, in fact, is to succeed in restoring some portion of
the accuracy of the associated forward solution, despite the fact that the
inverse problem, unlike the forward problem, is ill-posed. Thus it is
reasonable to validate an inverse solution with data that best matches the
forward solution. Based on validation studies with analytic solutions,[11] it is probably safe to assume that the errors that arise
in constructing a geometric model derived from a patient or animal play a
much larger role than those from the numerical methods used in the forward
solution. Additional errors such as movement during mechanical systole
simply compound the problem. By forward computing the torso potentials,
one can reduce--or at least control--errors due to geometry because these
errors are present in the forward solution. This strategy makes it
possible to focus the validation studies on the errors that arise in the
step of creating the inverse solution from the forward solution, which is
the major challenge in electrocardiographic inverse problems.
One weakness of this approach is that it neglects the effects of any errors
in the problem formulation, i.e., the equations used to describe the
relationship between sources and remote potentials. This omission occurs
because the same problem formulation is used in both the forward and
inverse solutions. There is a hybrid approach that uses dipole sources to
calculate both epicardial and torso surface potentials directly based on a
realistic geometry, but uses an explicit epicardial to torso surface forward
model, based on a different formulation, to generate separate
surface-to-surface forward and inverse solutions. Because the two
formulations are different, the dipole source forward model does not match
exactly the surface-to-surface model. One can then compare the directly
against the inversely computed epicardial potentials and perform cross
validation.[18,31] This model mismatch can reduce at
least some of the symmetry inherent in other computational approaches and
potentially provide less biased validation conditions, but, of course, is
limited to simple dipolar source models.
A more realistic hybrid approach is that described by Hren et al., in
which the source was not a dipole, but a cellular automata model of cardiac
propagation.[32,33,34] Cellular automata models
represent cardiac tissue as a regular mesh of ``cells'', each representing
a region of approximately
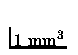 and use simplified cell-to-cell
coupling and state transition rules to determine the activation sequence.
Although not as detailed as the monodomain and bidomain models that are
based on descriptions of membrane kinetics, cellular automata models have a
long history in simulations of normal and abnormal cardiac
activation.[35,36,37,38,39,40,41,42,43,44,45]
In order to apply cellular automata models to validation of
electrocardiographic inverse solutions, Hren et al. developed a method
with which they assigned electrical source strength to the activation
wavefront and were thus able to compute epicardial and torso potentials
from the cellular automata model for realistic geometric models of the
human torso.[32,46] They used this technique to both
validate inverse solutions[33] and to examine the spatial
resolution of body surface mapping.[47,48]
and use simplified cell-to-cell
coupling and state transition rules to determine the activation sequence.
Although not as detailed as the monodomain and bidomain models that are
based on descriptions of membrane kinetics, cellular automata models have a
long history in simulations of normal and abnormal cardiac
activation.[35,36,37,38,39,40,41,42,43,44,45]
In order to apply cellular automata models to validation of
electrocardiographic inverse solutions, Hren et al. developed a method
with which they assigned electrical source strength to the activation
wavefront and were thus able to compute epicardial and torso potentials
from the cellular automata model for realistic geometric models of the
human torso.[32,46] They used this technique to both
validate inverse solutions[33] and to examine the spatial
resolution of body surface mapping.[47,48]
An obvious, significant strength of the computational and analytical
approaches is that they do not require the expense and extensive
infrastructure of experimental studies, but can all be performed on
a computer using methods that are well described in the literature.
Moreover, as the examination of the results leads to new hypotheses to
test, requiring new data, it is obviously easier to repeat a
computational experiment after modifying the conditions or changing
parameters than it is to have to modify and repeat experimental
studies.
Rob MacLeod
1999-11-06
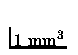 and use simplified cell-to-cell
coupling and state transition rules to determine the activation sequence.
Although not as detailed as the monodomain and bidomain models that are
based on descriptions of membrane kinetics, cellular automata models have a
long history in simulations of normal and abnormal cardiac
activation.[35,36,37,38,39,40,41,42,43,44,45]
In order to apply cellular automata models to validation of
electrocardiographic inverse solutions, Hren et al. developed a method
with which they assigned electrical source strength to the activation
wavefront and were thus able to compute epicardial and torso potentials
from the cellular automata model for realistic geometric models of the
human torso.[32,46] They used this technique to both
validate inverse solutions[33] and to examine the spatial
resolution of body surface mapping.[47,48]
and use simplified cell-to-cell
coupling and state transition rules to determine the activation sequence.
Although not as detailed as the monodomain and bidomain models that are
based on descriptions of membrane kinetics, cellular automata models have a
long history in simulations of normal and abnormal cardiac
activation.[35,36,37,38,39,40,41,42,43,44,45]
In order to apply cellular automata models to validation of
electrocardiographic inverse solutions, Hren et al. developed a method
with which they assigned electrical source strength to the activation
wavefront and were thus able to compute epicardial and torso potentials
from the cellular automata model for realistic geometric models of the
human torso.[32,46] They used this technique to both
validate inverse solutions[33] and to examine the spatial
resolution of body surface mapping.[47,48]