SCI Publications
2011


J.R. Anderson, S. Mohammed, B.C. Grimm, B.W. Jones, P. Koshevoy, T. Tasdizen, R.T. Whitaker, R.E. Marc.
“The Viking viewer for connectomics: scalable multi-user annotation and summarization of large volume data sets,” In Journal of Microscopy, Vol. 241, No. 1, pp. 13--28. 2011.
DOI: 10.1111/j.1365-2818.2010.03402.x
Modern microscope automation permits the collection of vast amounts of continuous anatomical imagery in both two and three dimensions. These large data sets present significant challenges for data storage, access, viewing, annotation and analysis. The cost and overhead of collecting and storing the data can be extremely high. Large data sets quickly exceed an individual's capability for timely analysis and present challenges in efficiently applying transforms, if needed. Finally annotated anatomical data sets can represent a significant investment of resources and should be easily accessible to the scientific community. The Viking application was our solution created to view and annotate a 16.5 TB ultrastructural retinal connectome volume and we demonstrate its utility in reconstructing neural networks for a distinctive retinal amacrine cell class. Viking has several key features. (1) It works over the internet using HTTP and supports many concurrent users limited only by hardware. (2) It supports a multi-user, collaborative annotation strategy. (3) It cleanly demarcates viewing and analysis from data collection and hosting. (4) It is capable of applying transformations in real-time. (5) It has an easily extensible user interface, allowing addition of specialized modules without rewriting the viewer.
Keywords: Annotation, automated electron microscopy, citizen science, computational methods, connectome, networks, visualization


M.L. Berlanga, S. Phan, E.A. Bushong, S. Lamont, S. Wu, O. Kwon, B.S. Phung, M. Terada, T. Tasdizen, E. Martone, M.H. Ellisman.
“Three-dimensional reconstruction of serial mouse brain sections using high-resolution large-scale mosaics,” In Frontiers in Neuroscience Methods, Vol. 5, pp. (published online). March, 2011.
DOI: 10.3389/fnana.2011.00017


L. Hogrebe, A. Paiva, E. Jurrus, C. Christensen, M. Bridge, J.R. Korenberg, T. Tasdizen.
“Trace Driven Registration of Neuron Confocal Microscopy Stacks,” In IEEE International Symposium on Biomedical Imaging (ISBI), pp. 1345--1348. 2011.
DOI: 10.1109/ISBI.2011.5872649


E. Jurrus, S. Watanabe, R. Guily, A.R.C. Paiva, M.H. Ellisman, E.M. Jorgensen, T. Tasdizen.
“Semi-automated Neuron Boundary Detection and Slice Traversal Algorithm for Segmentation of Neurons from Electron Microscopy Images,” In Microscopic Image Analysis with Applications in Biology (MIAAB) Workshop, 2011.
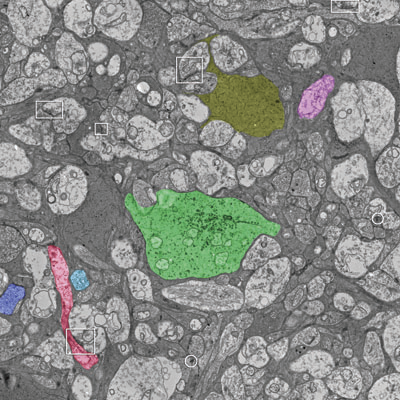
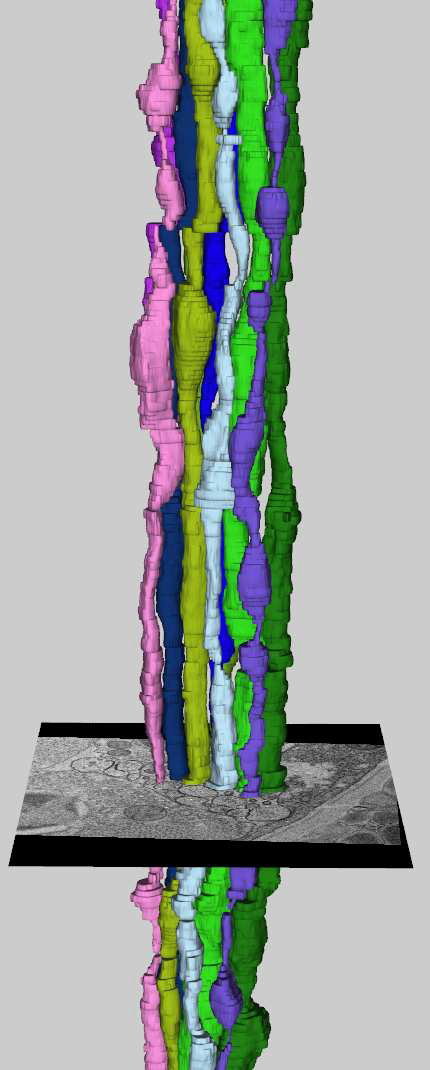
Neuroscientists are developing new imaging techniques and generating large volumes of data in an effort to understand the complex structure of the nervous system. To aid in the analysis, new segmentation techniques for identifying neurons in these feature rich datasets are required. However, the extremely anisotropic resolution of the data makes segmentation and tracking across slices difficult. This paper presents a complete method for segmenting neurons in electron microscopy images and visualizing them in three dimensions. First, we present an advanced method for identifying neuron membranes, necessary for whole neuron segmentation, using a machine learning approach. Next, neurons are segmented in each two-dimensional section and connected using correlation of regions between sections. These techniques, combined with a visual user interface, enable users to quickly segment whole neurons in large volumes.


Z. Leng, J.R. Korenberg, B. Roysam, T. Tasdizen.
“A rapid 2-D centerline extraction method based on tensor voting,” In 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 1000--1003. 2011.
DOI: 10.1109/ISBI.2011.5872570


M. Seyedhosseini, A.R.C. Paiva, T. Tasdizen.
“Multi-scale Series Contextual Model for Image Parsing,” SCI Technical Report, No. UUSCI-2011-004, SCI Institute, University of Utah, 2011.



M. Seyedhosseini, A.R.C. Paiva, T. Tasdizen.
“Fast AdaBoost training using weighted novelty selection,” In Proc. IEEE Intl. Joint Conf. on Neural Networks, San Jose, CA, USA pp. 1245--1250. August, 2011.
In this paper, a new AdaBoost learning framework, called WNS-AdaBoost, is proposed for training discriminative models. The proposed approach significantly speeds up the learning process of adaptive boosting (AdaBoost) by reducing the number of data points. For this purpose, we introduce the weighted novelty selection (WNS) sampling strategy and combine it with AdaBoost to obtain an efficient and fast learning algorithm. WNS selects a representative subset of data thereby reducing the number of data points onto which AdaBoost is applied. In addition, WNS associates a weight with each selected data point such that the weighted subset approximates the distribution of all the training data. This ensures that AdaBoost can trained efficiently and with minimal loss of accuracy. The performance of WNS-AdaBoost is first demonstrated in a classification task. Then, WNS is employed in a probabilistic boosting-tree (PBT) structure for image segmentation. Results in these two applications show that the training time using WNS-AdaBoost is greatly reduced at the cost of only a few percent in accuracy.


M. Seyedhosseini, R. Kumar, E. Jurrus, R. Guily, M. Ellisman, H. Pfister, T. Tasdizen.
“Detection of Neuron Membranes in Electron Microscopy Images using Multi-scale Context and Radon-like Features,” In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2011, Lecture Notes in Computer Science (LNCS), Vol. 6891, pp. 670--677. 2011.
DOI: 10.1007/978-3-642-23623-5_84
2010


G. Adluru, T. Tasdizen, M.C. Schabel, E.V.R. DiBella.
“Reconstruction of 3D Dynamic Contrast-Enhanced Magnetic Resonance Imaging Using Nonlocal Means,” In Journal of Magnetic Resonance Imaging, Vol. 32, pp. 1217--1227. 2010.
DOI: 10.1002/jmri.22358


G. Adluru, T. Tasdizen, R. Whitaker, E. DiBella.
“Improving Undersampled MRI Reconstruction Using Non-Local Means,” In Proceedings of the 2010 International Conference on Pattern Recognition, pp. 4000--4003. 2010.
DOI: 10.1109/ICPR.2010.973


J.R. Anderson, B.C. Grimm, S. Mohammed, B.W. Jones, T. Tasdizen, J. Spaltenstein, P. Koshevoy, R.T. Whitaker, R.E. Marc.
“The Viking Viewer: Scalable Multiuser Annotation and Summarization of Large Volume Datasets,” In Journal of Microscopy, Vol. 241, No. 1, pp. 13--28. 2010.
DOI: 10.1111/j.1365-2818.2010.03402.x


S. Gerber, T. Tasdizen, P.T. Fletcher, S. Joshi, R.T. Whitaker, the Alzheimers Disease Neuroimaging Initiative (ADNI).
“Manifold modeling for brain population analysis,” In Medical Image Analysis, Special Issue on the 12th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2009, Vol. 14, No. 5, Note: Awarded MICCAI 2010, Best of the Journal Issue Award, pp. 643--653. 2010.
ISSN: 1361-8415
DOI: 10.1016/j.media.2010.05.008
PubMed ID: 20579930


S.K. Iyer, E. DiBella, T. Tasdizen.
“Edge enhanced spatio-temporal constrained reconstruction of undersampled dynamic contrast enhanced radial MRI,” In IEEE International Symposium on Biomedical Imaging (ISBI): From Nano to Macro, pp. 704--707. 2010.
DOI: 10.1109/ISBI.2010.5490077


E. Jurrus, A.R.C. Paiva, S. Watanabe, J.R. Anderson, B.W. Jones, R.T. Whitaker, E.M. Jorgensen, R.E. Marc, T. Tasdizen.
“Detection of Neuron Membranes in Electron Microscopy Images Using a Serial Neural Network Architecture,” In Medical Image Analysis, Vol. 14, No. 6, pp. 770--783. 2010.
DOI: 10.1016/j.media.2010.06.002
PubMed ID: 20598935


A.R.C. Paiva, T. Tasdizen.
“Fast Semi-Supervised Image Segmentation by Novelty Selection,” In Proceedings of IEEE International Conference on Acoustics, Speech and Signal Processing, Dallas, Texas, pp. 1054--1057. March, 2010.
DOI: 10.1109/ICASSP.2010.5495333


A.R.C. Paiva, T. Tasdizen.
“Detection of Salient Image Points using Manifold Structure,” In Proc. IEEE Intl. Conference on Pattern Recognition, Istanbul, Turkey, pp. 1389--1392. 2010.
DOI: 10.1109/ICPR.2010.343


A.R.C. Paiva, E. Jurrus, T. Tasdizen.
“Using Sequential Context for Image Analysis,” In Proc. IEEE Intl. Conference on Pattern Recognition, Istanbul, Turkey, pp. 2800--2803. 2010.
DOI: 10.1109/ICPR.2010.686


C. Schlimper, O. Nemitz, U. Dorenbeck, J. Scorzin, R.T. Whitaker, T. Tasdizen, M. Rumpf, K. Schaller.
“Restoring three-dimensional magnetic resonance angiography images with mean curvature motion,” In Neurological Research, Vol. 32, No. 1, Note: Epub 2009 Nov 26, pp. 87--93. February, 2010.
DOI: 10.1179/016164110X12556180206077
PubMed ID: 19941735


M. Seyedhosseini, A.R.C. Paiva, T. Tasdizen.
“Image Parsing with a Three-State Series Neural Network Classifier,” In Proc. IEEE Intl. Conference on Pattern Recognition, Istanbul, Turkey, pp. 4508--4511. 2010.
DOI: 10.1109/ICPR.2010.1095


T. Tasdizen, P. Koshevoy, B.C. Grimm, J.R. Anderson, B.W. Jones, C.B. Watt, R.T. Whitaker, R.E. Marc.
“Automatic mosaicking and volume assembly for high-throughput serial-section transmission electron microscopy,” In Journal of Neuroscience Methods, Vol. 193, No. 1, pp. 132--144. 2010.
PubMed ID: 20713087