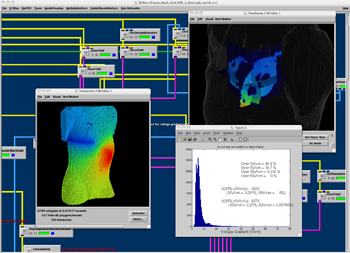
 BioPSE network computing the effects of electrode placement on the electrical field in the heart and hence evaluating the defibrillation threshold. |
The Biomedical Problem Solving Environment, or BioPSE, contains a suite of applications and integrated problem solving systems that have been specialized to meet the needs of scientists working in major fields of biomedicine. In order to design the best systems to meet real world needs, the CIBC has established collaborations with prominent scientists who are leaders in their fields. The goal of these collaborations is to develop software that is well suited to the needs of scientists working in these and related fields of biomedicine.
One of the real world problems guiding the development of BioPSE is the need to predict optimal placement of Implantable Cardiac Defibrillators (ICDs) in pediatric patients. Young patients present a unique challenge due to the natural anatomical and physiological differences found in newborns to adolescents. Predicting how current flow will be affected by electrode placement and interaction with an individual's anatomy is a complex but important part of preoperative planning. Electrode placement is crucial in order to deliver the right amount of current to the heart for defibrillation without damaging any tissues.
BioPSE developers at the CIBC are working in collaboration with Dr. John Triedman and Dr. Matthew Jolley at Children's Hospital in Boston to develop tools for generating custom computer models of individual patients and simulating the activity of implanted cardiac defibrillators in their torsos. BioPSE provides an intuitive three-dimensional simulation environment with which doctors can move the electrodes around and immediately see the resulting currents that would be delivered to the heart. This instant visual feedback allows the doctors to locate the ideal placement for the electrodes before cutting into a patient.
"We utilize SCIRun and BioPSE to interactively explore novel locations for implantable cardiac defibrillators (ICDs) in children. ICD electrode placement in kids is difficult due to their size and other anatomical limitations. As a result, many children require a unique placement performed on an ad hoc basis. SCIRun and BioPSE provide a easy to use environment to place virtual electrodes in individual child torso models and subsequently compute and visualize the expected defibrillating electric fields, allowing logical placement of electrodes rather than clinical trial and error. Our research would not be possible without the ongoing development and customization of these tools by the SCI Institute." -- Dr. Matthew Jolley, Department of Cardiology, Children's Hospital Boston.
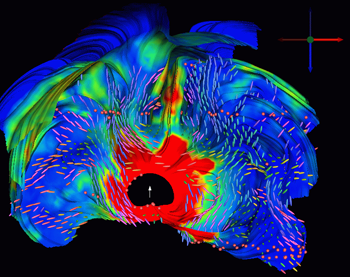 Visualization of the return current tensors for a deep thalamic source in volume conductor with anisotropic white matter compartment. |
"We use BioPSE in Electroencephalography (EEG) and Magnetoencephalography (MEG) source analysis for visualization purposes. BioPSE allows us to visualize and thus validate MRI-based head tissue segmentation, and tissue conductivity tensor labeling, as well as field distributions in complicated volume conductor models of the human head. These scientific visualization tasks are indispensable for a successful source analysis in areas like epilepsy or more generally in the area of the neurosciences." -- Dr. Carsten Wolters, Institute for Biomagnetism and Biosignal Analysis, University of Muenster, Germany.These are just two example applications of this highly adaptable and extensible biomedical problem solving environment. BioPSE achieves this adaptability though the dataflow paradigm. This breaks any scientific problem into a series of steps which are handled by modules. Data flows from one module to the next, each solving an aspect of the problem, until it reaches the last module, typically the viewer, which outputs the results of the process to the screen. Each module comes with an intuitive user interface for adjusting the parameters relevant to its part of the process. BioPSE comes with a library of these modules and pre-built networks for common biomedical applications. New users can rely on the pre-built networks to handle most common tasks, while more experienced users can construct their own networks or even create their own custom modules. BioPSE also comes with a number of PowerApps, or expansion packages which extend its capabilities for specific applications. CIBC developers and members of the BioPSE community of users are continually developing more tools and applications for BioPSE. Many of the new features available in SCIRun/BioPSE 3.0 are a direct result of the close work we've done with our collaborators. For example, a new PowerApp called Seg3D was added to address our collaborators' need for automatic segmentation of three-dimensional medical imaging data.
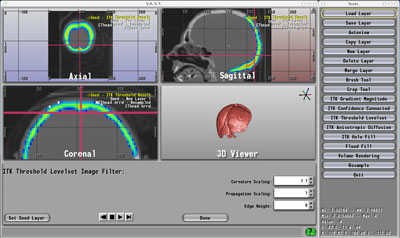 Seg 3D |
"We developed Seg3D mainly in response to requests from Mark Ellisman's National Center for Microscopy and Imaging Research (NCMIR) group at UCSD. Specifically, they asked for a 3D segmentation tool that will accommodate manual segmentations and respect ontological information, but will also allow them to experiment with a variety of automated algorithms, such as those found in ITK." -- Joshua Cates, BioPSE Developer, Scientific Computing and Imaging Institute, University of Utah.The next major release of BioPSE, expected in the Summer of 2007, will include a number of new features such as additional tools for meshing.
Please visit the CIBC Software website to download SCIRun/BioPSE 3.0. Installation and documentation are now available on our SCIRun Documentation Wiki.
