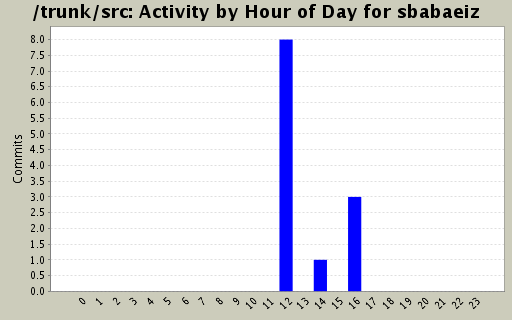
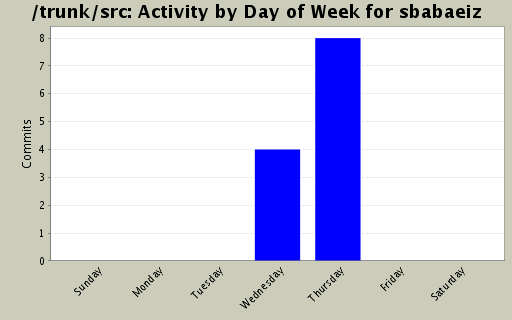
Activity by Clock Time


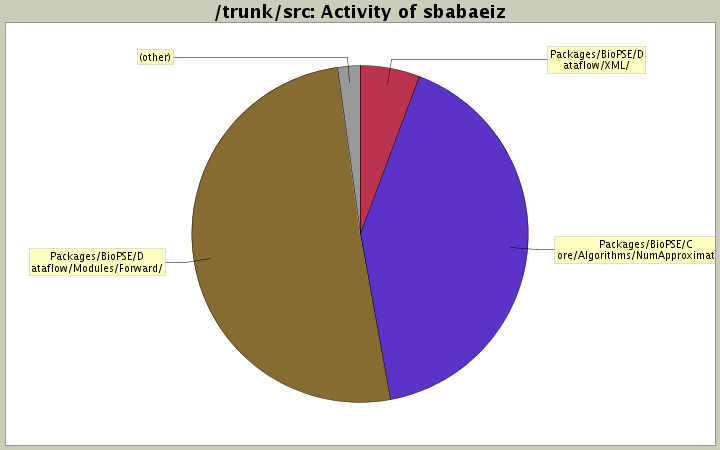
| Directory | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 12 (100.0%) | 1941 (100.0%) | 161.7 |
| Packages/BioPSE/Dataflow/Modules/Forward/ | 5 (41.7%) | 985 (50.7%) | 197.0 |
| Packages/BioPSE/Core/Algorithms/NumApproximation/ | 4 (33.3%) | 802 (41.3%) | 200.5 |
| Packages/BioPSE/Dataflow/XML/ | 1 (8.3%) | 112 (5.8%) | 112.0 |
| Core/Datatypes/ | 2 (16.7%) | 42 (2.2%) | 21.0 |
For SetupEITMatrix module
112 lines of code changed in 1 file:
Solved the forward problem of EIT
529 lines of code changed in 2 files:
adding SetupEITMatrix which solves the forward problem of EIT
2 lines of code changed in 1 file:
Instead of adding the module I had developed for Eric Voth to solve the BEM-ESI problem to SCIRun, I modified the existing SetupBEMatrix.cc, so now it works for both BEM-ECG and BEM-ESI problems.
454 lines of code changed in 2 files:
I added the general BEM matrices to a new class BuildBEMatrix (cc and h) in /src/Packages/BioPSE/Core/Algorithms/NumApproximation/ and added BuildBEMatrix.cc file in the corresponding sub.mk file. Then changed SetupBEMatrix.h and SetupBEMatrix.cc accordingly
802 lines of code changed in 2 files:
I added the general BEM matrices to a new class BuildBEMatrix (cc and h) in /src/Packages/BioPSE/Core/Algorithms/NumApproximation/ and added BuildBEMatrix.cc file in the corresponding sub.mk file. Then changed SetupBEMatrix.h and SetupBEMatrix.cc accordingly
0 lines of code changed in 2 files:
I added two functions Concat_row() and Concat_cols() to DenseMatrix class as Friend functions: (lines 160 and 161 in DenseMatrix.h , lines 1120-1155 in DenseMatrix.cc)
42 lines of code changed in 2 files: