Parallel I/O Framework
PIDX is a scalable and tunable adaptive resolution parallel I/O framework
Overview
Deployments
Tutorials
Team
Publications
PIDX
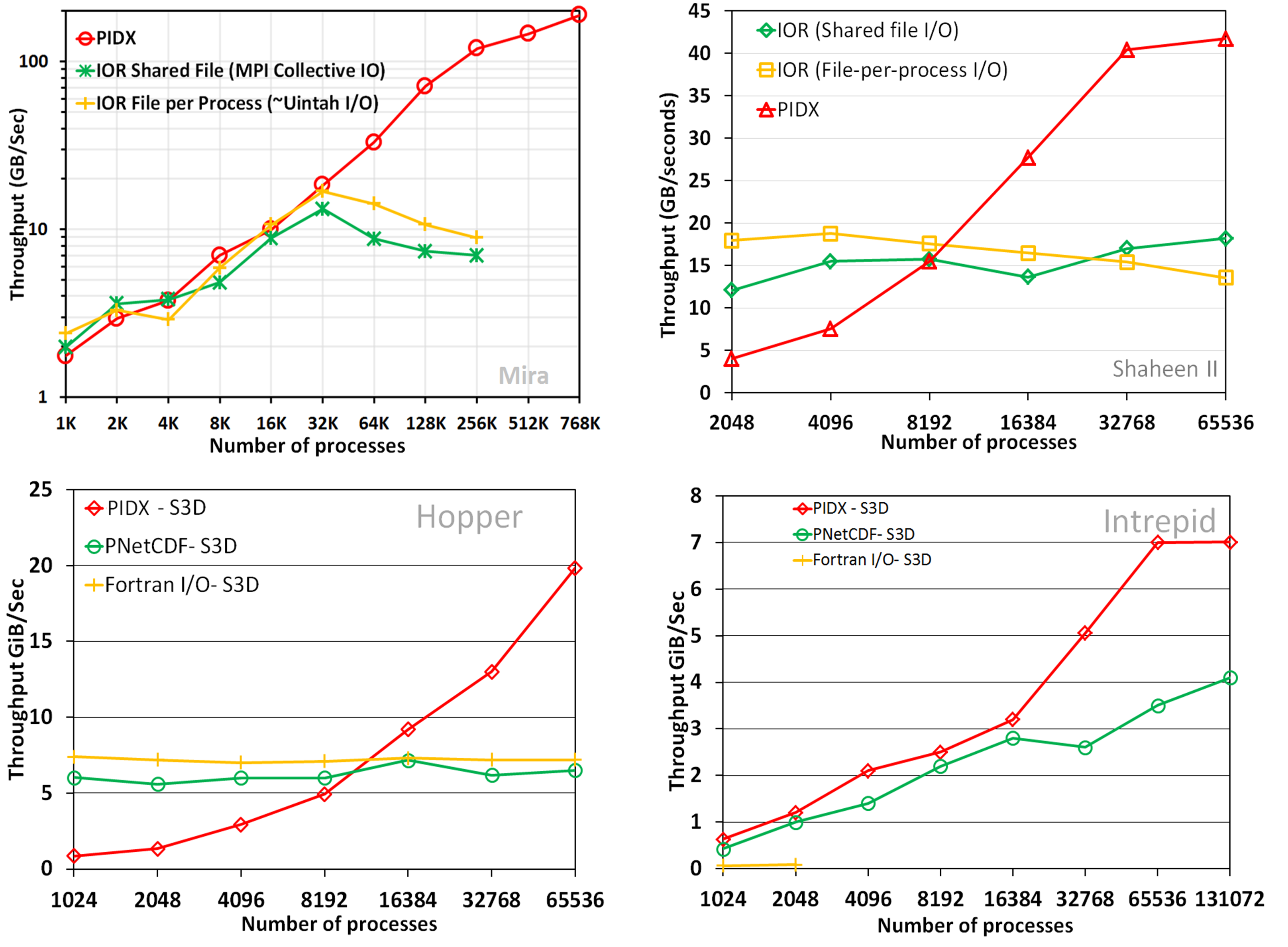
The increase in computational power of supercomputers is enabling complex scientific phenomena to be simulated at ever-increasing resolution and fidelity. With these simulations routinely producing large volumes of data, performing efficient I/O at this scale has become a very difficult task. Large-scale parallel writes are challenging due to the complex interdependencies between I/O middleware and hardware. Analytic-appropriate reads are traditionally hindered by bottlenecks in I/O access. Moreover, the two components of I/O, data generation from simulations (writes) and data exploration for analysis and visualization (reads), have substantially different data access requirements. Parallel writes, performed on supercomputers, often deploy aggregation strategies to permit large-sized contiguous access. Analysis and visualization tasks, usually performed on computationally modest resources, require fast access to localized subsets or multiresolution representations of the data. This work tackles the problem of parallel I/O while bridging the gap between large-scale writes and analytics-appropriate reads.The focus of this work is to develop an end-to-end adaptive-resolution data movement framework that provides efficient I/O, while supporting the full spectrum of modern HPC hardware. This is achieved by developing technology for highly scalable and tunable parallel I/O, applicable to both traditional parallel data formats and multiresolution data formats, which are directly appropriate for analysis and visualization. To demonstrate the efficacy of the approach, a novel library, PIDX is developed that is highly tunable and capable of adaptive-resolution parallel I/O to a multiresolution data format. Adaptive resolution storage and I/O, which allows subsets of a simulation to be accessed at varying spatial resolutions, can yield significant improvements to both the storage performance and I/O time. The library provides a set of parameters that controls the storage format and the nature of data aggregation across the network; further, a machine learning-based model is constructed that tunes these parameters for the maximum throughput. This work is empirically demonstrated by showing parallel I/O scaling up to 768K cores within a framework flexible enough to handle adaptive resolution I/O.
The Team
 |
 |
 |
 |
 |
| Sidharth Kumar | Steve Petruzza | Duong hoang | Cameron Christenson | Valerio Pascucci |
Publications
S. Kumar, C. Christensen, P.-T. Bremer, E. Brugger, V. Pascucci, J. Schmidt, M. Berzins, H. Kolla, J. Chen, V. Vishwanath, P. Carns, R. Grout. “Fast Multi-Resolution Reads of Massive Simulation Datasets,” In Proceedings of the International Supercomputing Conference ISC'14, Leipzig, Germany, June, 2014.S. Kumar, J. Edwards, P.-T. Bremer, A. Knoll, C. Christensen, V. Vishwanath, P. Carns, J.A. Schmidt, V. Pascucci. “Efficient I/O and storage of adaptive-resolution data,” In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, IEEE Press, pp. 413--423. 2014.
S. Kumar, A. Saha, V. Vishwanath, P. Carns, J.A. Schmidt, G. Scorzelli, H. Kolla, R. Grout, R. Latham, R. Ross, M.E. Papka, J. Chen, V. Pascucci. “Characterization and modeling of PIDX parallel I/O for performance optimization,” In Proceedings of SC13: International Conference for High Performance Computing, Networking, Storage and Analysis, pp. 67. 2013.
V. Pascucci, P.-T. Bremer, A. Gyulassy, G. Scorzelli, C. Christensen, B. Summa, S. Kumar. “Scalable Visualization and Interactive Analysis Using Massive Data Streams,” In Cloud Computing and Big Data, Advances in Parallel Computing, Vol. 23, IOS Press, pp. 212--230. 2013.
S. Kumar, V. Vishwanath, P. Carns, J.A. Levine, R. Latham, G. Scorzelli, H. Kolla, R. Grout, R. Ross, M.E. Papka, J. Chen, V. Pascucci. “Efficient data restructuring and aggregation for I/O acceleration in PIDX,” In Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, IEEE Computer Society Press, pp. 50:1--50:11. 2012.
V. Pascucci, G. Scorzelli, B. Summa, P.-T. Bremer, A. Gyulassy, C. Christensen, S. Philip, S. Kumar. “The ViSUS Visualization Framework,” In High Performance Visualization: Enabling Extreme-Scale Scientific Insight, Chapman and Hall/CRC Computational Science, Ch. 19, Edited by E. Wes Bethel and Hank Childs (LBNL) and Charles Hansen (UofU), Chapman and Hall/CRC, 2012.
S. Kumar, V. Vishwanath, P. Carns, B. Summa, G. Scorzelli, V. Pascucci, R. Ross, J. Chen, H. Kolla, R. Grout. “PIDX: Efficient Parallel I/O for Multi-resolution Multi-dimensional Scientific Datasets,” In Proceedings of The IEEE International Conference on Cluster Computing, pp. 103--111. September, 2011.
S. Kumar, V. Vishwanath, P. Carns, V. Pascucci, R. Latham, T. Peterka, M. Papka, R. Ross. “Towards Efficient Access of Multi-dimensional, Multi-resolution Scientific Data,” In Proceedings of the 5th Petascale Data Storage Workshop, Supercomputing 2010, pp. (in press). 2010.