SCI Publications
2012
Y. Hong, S. Joshi, M. Sanchez, M. Styner, M. Niethammer.
“Metamorphic Geodesic Regression,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2012, pp. 197--205. 2012.
V. Sarkar, Brian Wang, J. Hinkle, V.J. Gonzalez, Y.J. Hitchcock, P. Rassiah-Szegedi, S. Joshi, B.J. Salter.
“Dosimetric evaluation of a virtual image-guidance alternative to explicit 6 degree of freedom robotic couch correction,” In Practical Radiation Oncology, Vol. 2, No. 2, pp. 122--137. 2012.
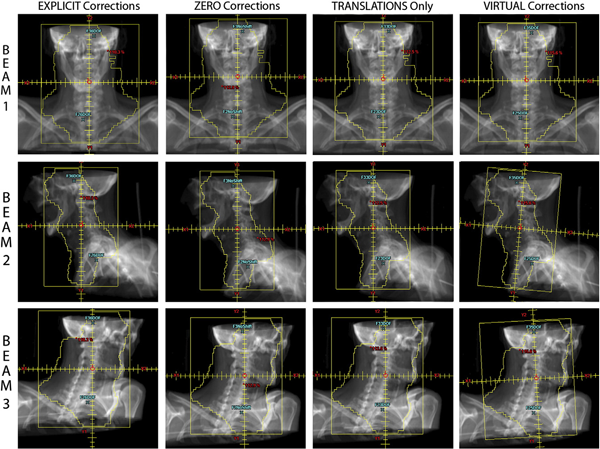
Purpose: Clinical evaluation of a \"virtual\" methodology for providing 6 degrees of freedom (6DOF) patient set-up corrections and comparison to corrections facilitated by a 6DOF robotic couch.
Methods: A total of 55 weekly in-room image-guidance computed tomographic (CT) scans were acquired using a CT-on-rails for 11 pelvic and head and neck cancer patients treated at our facility. Fusion of the CT-of-the-day to the simulation CT allowed prototype virtual 6DOF correction software to calculate the translations, single couch yaw, and beam-specific gantry and collimator rotations necessary to effectively reproduce the same corrections as a 6DOF robotic couch. These corrections were then used to modify the original treatment plan beam geometry and this modified plan geometry was applied to the CT-of-the-day to evaluate the dosimetric effects of the virtual correction method. This virtual correction dosimetry was compared with calculated geometric and dosimetric results for an explicit 6DOF robotic couch correction methodology.
Results: A (2\%, 2mm) gamma analysis comparing dose distributions created using the virtual corrections to those from explicit corrections showed that an average of 95.1\% of all points had a gamma of 1 or less, with a standard deviation of 3.4\%. For a total of 470 dosimetric metrics (ie, maximum and mean dose statistics for all relevant structures) compared for all 55 image-guidance sessions, the average dose difference for these metrics between the plans employing the virtual corrections and the explicit corrections was -0.12\% with a standard deviation of 0.82\%; 97.9\% of all metrics were within 2\%.
Conclusions: Results showed that the virtual corrections yielded dosimetric distributions that were essentially equivalent to those obtained when 6DOF robotic corrections were used, and that always outperformed the most commonly employed clinical approach of 3 translations only. This suggests
N.P. Singh, A.Y. Wang, P. Sankaranarayanan, P.T. Fletcher, S. Joshi.
“Genetic, Structural and Functional Imaging Biomarkers for Early Detection of Conversion from MCI to AD,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention MICCAI 2012, Vol. 7510, pp. 132--140. 2012.
DOI: 10.1007/978-3-642-33415-3_17
With the advent of advanced imaging techniques, genotyping, and methods to assess clinical and biological progression, there is a growing need for a unified framework that could exploit information available from multiple sources to aid diagnosis and the identification of early signs of Alzheimer’s disease (AD). We propose a modeling strategy using supervised feature extraction to optimally combine highdimensional imaging modalities with several other low-dimensional disease risk factors. The motivation is to discover new imaging biomarkers and use them in conjunction with other known biomarkers for prognosis of individuals at high risk of developing AD. Our framework also has the ability to assess the relative importance of imaging modalities for predicting AD conversion. We evaluate the proposed methodology on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database to predict conversion of individuals with Mild Cognitive Impairment (MCI) to AD, only using information available at baseline.
Keywords: adni
M. Szegedi, P. Rassiah-Szegedi, V. Sarkar, J. Hinkle, Brian Wang, Y.-H. Huang, H. Zhao, S. Joshi, B.J. Salter.
“Tissue characterization using a phantom to validate four-dimensional tissue deformation,” In Medical Physics, Vol. 39, No. 10, pp. 6065--6070. 2012.
DOI: 10.1118/1.4747528
Purpose: This project proposes using a real tissue phantom for 4D tissue deformation reconstruction (4DTDR) and 4D deformable image registration (DIR) validation, which allows for the complete verification of the motion path rather than limited end-point to end-point of motion.
Methods: Three electro-magnetic-tracking (EMT) fiducials were implanted into fresh porcine liver that was subsequently animated in a clinically realistic phantom. The animation was previously shown to be similar to organ motion, including hysteresis, when driven using a real patient's breathing pattern. For this experiment, 4DCTs and EMT traces were acquired when the phantom was animated using both sinusoidal and recorded patient-breathing traces. Fiducial were masked prior to 4DTDR for reconstruction. The original 4DCT data (with fiducials) were sampled into 20 CT phase sets and fiducials’ coordinates were recorded, resulting in time-resolved fiducial motion paths. Measured values of fiducial location were compared to EMT measured traces and the result calculated by 4DTDR.
Results: For the sinusoidal breathing trace, 95\% of EMT measured locations were within 1.2 mm of the measured 4DCT motion path, allowing for repeatable accurate motion characterization. The 4DTDR traces matched 95\% of the EMT trace within 1.6 mm. Using the more irregular (in amplitude and frequency) patient trace, 95\% of the EMT trace points fitted both 4DCT and 4DTDR motion path within 4.5 mm. The average match of the 4DTDR estimation of the tissue hysteresis over all CT phases was 0.9 mm using a sinusoidal signal for animation and 1.0 mm using the patient trace.
Conclusions: The real tissue phantom is a tool which can be used to accurately characterize tissue deformation, helping to validate or evaluate a DIR or 4DTDR algorithm over a complete motion path. The phantom is capable of validating, evaluating, and quantifying tissue hysteresis, thereby allowing for full motion path validation.
2011
S. Durrleman, M.W. Prastawa, G. Gerig, S. Joshi.
“Optimal data-driven sparse parameterization of diffeomorphisms for population analysis,” In Proceedings of the IPMI 2011 conference, Springer LNCS, Vol. 6801/2011, pp. 123--134. July, 2011.
DOI: 10.1007/978-3-642-22092-0_11
PubMed ID: 20516153
S.E. Geneser, J.D. Hinkle, R.M. Kirby, Bo Wang, B. Salter, S. Joshi.
“Quantifying variability in radiation dose due to respiratory-induced tumor motion,” In Medical Image Analysis, Vol. 15, No. 4, pp. 640--649. 2011.
DOI: 10.1016/j.media.2010.07.003
L.K. Ha, J. Krüger, J. Comba, S. Joshi, C.T. Silva.
“Optimal Multi-Image Processing Streaming Framework on Parallel Heterogeneous Systems,” In Proceedings of Eurographics Symposium on Parallel Graphics and Visualization 2011, Note: Awarded Best Paper!, pp. 1--10. 2011.
DOI: 10.2312/EGPGV/EGPGV11/001-010
Atlas construction is an important technique in medical image analysis that plays a central role in understanding the variability of brain anatomy. The construction often requires applying image processing operations to multiple images (often hundreds of volumetric datasets), which is challenging in computational power as well as memory requirements. In this paper we introduce MIP, a Multi-Image Processing streaming framework to harness the processing power of heterogeneous CPU/GPU systems. In MIP we introduce specially designed streaming algorithms and data structures that provides an optimal solution for out-of-core multi-image processing problems both in terms of memory usage and computational efficiency. MIP makes use of the asynchronous execution mechanism supported by parallel heterogeneous systems to efficiently hide the inherent latency of the processing pipeline of out-of-core approaches. Consequently, with computationally intensive problems, the MIP out-of-core solution could achieve the same performance as the in-core solution. We demonstrate the efficiency of the MIP framework on synthetic and real datasets.
L.K. Ha, J. Krüger, S. Joshi, C.T. Silva.
“Multi-scale Unbiased Diffeomorphic Atlas Construction on Multi-GPUs,” Vol. 1, Ch. 10, Morgan Kaufmann, pp. 42. 2011.
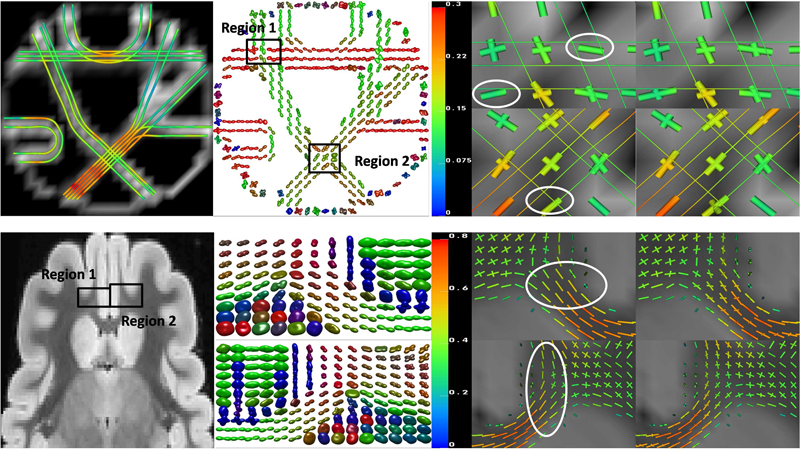
F. Jiao, Y. Gur, C.R. Johnson, S. Joshi.
“Detection of crossing white matter fibers with high-order tensors and rank-k decompositions,” In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI 2011), Lecture Notes in Computer Science (LNCS), Vol. 6801, pp. 538--549. 2011.
DOI: 10.1007/978-3-642-22092-0_44
PubMed Central ID: PMC3327305
B. Salter, B. Wang, M. Sadinski, S. Ruhnau, V. Sarkar, J. Hinkle, Y. Hitchcock, K. Kokeny, S. Joshi.
“WE-E-BRC-06: Comparison of Two Methods of Contouring Internal Target Volume on Multiple 4DCT Data Sets from the Same Subjects: Maximum Intensity Projection and Combination of 10 Phases,” In Medical Physics, Vol. 38, No. 6, pp. 3820. 2011.
M. Szegedi, J. Hinkle, S. Joshi, V. Sarkar, P. Rassiah-Szegedi, B. Wang, B. Salter.
“WE-E-BRC-05: Voxel Based Four Dimensional Tissue Deformation Reconstruction (4DTDR) Validation Using a Real Tissue Phantom,” In Medical Physics, Vol. 38, pp. 3819. 2011.
2010
J.J.E. Blauer, J. Cates, C.J. McGann, E.G. Kholmovski, A. Alexander, M.W. Prastawa, S. Joshi, N.F. Marrouche, R.S. MacLeod.
“MRI Based Injury Characterization Immediately Following Ablation of Atrial Fibrillation,” In Computing in Cardiology, Vol. 37, pp. 165--168. 2010.
ISSN: 0276−6574
B.C. Davis, P.T. Fletcher, E. Bullitt, S. Joshi.
“Population Shape Regression from Random Design Data,” In International Journal of Computer Vision, Vol. 90, No. 1, Note: Marr Prize Special Issue, pp. 255--266. October, 2010.
DOI: 10.1109/ICCV.2007.4408977
S.E. Geneser, J.D. Hinkle, R.M. Kirby, Brian Wang, B. Salter, S. Joshi.
“Quantifying Variability in Radiation Dose Due to Respiratory-Induced Tumor Motion,” In Medical Image Analysis, Vol. 15, No. 4, pp. 640--649. 2010.
DOI: 10.1016/j.media.2010.07.003
S. Gerber, T. Tasdizen, P.T. Fletcher, S. Joshi, R.T. Whitaker, the Alzheimers Disease Neuroimaging Initiative (ADNI).
“Manifold modeling for brain population analysis,” In Medical Image Analysis, Special Issue on the 12th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2009, Vol. 14, No. 5, Note: Awarded MICCAI 2010, Best of the Journal Issue Award, pp. 643--653. 2010.
ISSN: 1361-8415
DOI: 10.1016/j.media.2010.05.008
PubMed ID: 20579930
L. Ha, M.W. Prastawa, G. Gerig, J.H. Gilmore, C.T. Silva, S. Joshi.
“Image Registration Driven by Combined Probabilistic and Geometric Descriptors,” In Med Image Comput Comput Assist Interv., Vol. 13, No. 2, pp. 602--609. 2010.
PubMed ID: 20879365
L.K. Ha, J. Krüger, S. Joshi, C.T. Silva.
“Multi-scale Unbiased Diffeomorphic Atlas Construction on Multi-GPUs,” In GPU Computing Gems, Vol. 1, 2010.
In this chapter, we present a high performance multi-scale 3D image processing framework to exploit the parallel processing power of multiple graphic processing units (Multi-GPUs) for medical image analysis. We developed GPU algorithms and data structures that can be applied to a wide range of 3D image processing applications and efficiently exploit the computational power and massive bandwidth offered by modern GPUs. Our framework helps scientists solve computationally intensive problems which previously required super computing power. To demonstrate the effectiveness of our framework and to compare to existing techniques, we focus our discussions on atlas construction - the application of understanding the development of the brain and the progression of brain diseases.
L.K. Ha, M.W. Prastawa, G. Gerig, J.H. Gilmore, C.T. Silva, S. Joshi.
“Image Registration Driven by Combined Probabilistic and Geometric Descriptors,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention – MICCAI 2010, Lecture Notes in Computer Science (LNCS), Vol. 6362/2010, pp. 602--609. 2010.
DOI: 10.1007/978-3-642-15745-5_74
Deformable image registration in the presence of considerable contrast differences and large-scale size and shape changes represents a significant challenge for image registration. A representative driving application is the study of early brain development in neuroimaging, which requires co-registration of images of the same subject across time or building 4-D population atlases. Growth during the first few years of development involves significant changes in size and shape of anatomical structures but also rapid changes in tissue properties due to myelination and structuring that are reflected in the multi-modal Magnetic Resonance (MR) contrastmeasurements. We propose a new registration method that generates a mapping between brain anatomies represented as a multi-compartment model of tissue class posterior images and geometries.We transform intensity patterns into combined probabilistic and geometric descriptors that drive thematching in a diffeomorphic framework, where distances between geometries are represented using currents which does not require geometric correspondence. We show preliminary results on the registrations of neonatal brainMRIs to two-year old infantMRIs using class posteriors and surface boundaries of structures undergoing major changes. Quantitative validation demonstrates that our proposedmethod generates registrations that better preserve the consistency of anatomical structures over time.
Keywords: netl
N.P. Singh, P.T. Fletcher, J.S. Preston, L. Ha, R. King, J.S. Marron, M. Wiener, S. Joshi.
“Multivariate Statistical Analysis of Deformation Momenta Relating Anatomical Shape to Neuropsychological Measures,” In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2010, Lecture Notes in Computer Science (LCNS), Vol. 6363/2010, pp. 529-537. 2010.
DOI: 10.1007/978-3-642-15711-0_66
PubMed ID: 20879441
2009
P.T. Fletcher PT, S. Venkatasubramanian, S. Joshi.
“The geometric median on Riemannian manifolds with application to robust atlas estimation,” In Neuroimage, Vol. 45, No. 1, pp. S143--S152. March, 2009.
PubMed ID: 19056498
Page 3 of 7
