SCI Publications
2016
S.K. Iyer, T. Tasdizen, N. Burgon, E. Kholmovski, N. Marrouche, G. Adluru, E.V.R. DiBella.
“Compressed sending for rapid late gadolinium enhanced imaging of the left atrium: A preliminary study, Magnetic Resonance Imaging,” In Magnetic Resonance Imaging, Vol. 34, No. 7, Elsevier BV, pp. 846--854. September, 2016.
DOI: 10.1016/j.mri.2016.03.002
Current late gadolinium enhancement (LGE) imaging of left atrial (LA) scar or fibrosis is relatively slow and requires 5–15 min to acquire an undersampled (R = 1.7) 3D navigated dataset. The GeneRalized Autocalibrating Partially Parallel Acquisitions (GRAPPA) based parallel imaging method is the current clinical standard for accelerating 3D LGE imaging of the LA and permits an acceleration factor ~ R = 1.7. Two compressed sensing (CS) methods have been developed to achieve higher acceleration factors: a patch based collaborative filtering technique tested with acceleration factor R ~ 3, and a technique that uses a 3D radial stack-of-stars acquisition pattern (R ~ 1.8) with a 3D total variation constraint. The long reconstruction time of these CS methods makes them unwieldy to use, especially the patch based collaborative filtering technique. In addition, the effect of CS techniques on the quantification of percentage of scar/fibrosis is not known.
We sought to develop a practical compressed sensing method for imaging the LA at high acceleration factors. In order to develop a clinically viable method with short reconstruction time, a Split Bregman (SB) reconstruction method with 3D total variation (TV) constraints was developed and implemented. The method was tested on 8 atrial fibrillation patients (4 pre-ablation and 4 post-ablation datasets). Blur metric, normalized mean squared error and peak signal to noise ratio were used as metrics to analyze the quality of the reconstructed images, Quantification of the extent of LGE was performed on the undersampled images and compared with the fully sampled images. Quantification of scar from post-ablation datasets and quantification of fibrosis from pre-ablation datasets showed that acceleration factors up to R ~ 3.5 gave good 3D LGE images of the LA wall, using a 3D TV constraint and constrained SB methods. This corresponds to reducing the scan time by half, compared to currently used GRAPPA methods. Reconstruction of 3D LGE images using the SB method was over 20 times faster than standard gradient descent methods.
T. Liu, S.M. Seyedhosseini, T. Tasdizen.
“Image Segmentation Using Hierarchical Merge Tree,” In IEEE Transactions on Image Processing, Vol. 25, No. 10, IEEE, pp. 4596--4607. Oct, 2016.
DOI: 10.1109/tip.2016.2592704
This paper investigates one of the most fundamental computer vision problems: image segmentation. We propose a supervised hierarchical approach to object-independent image segmentation. Starting with oversegmenting superpixels, we use a tree structure to represent the hierarchy of region merging, by which we reduce the problem of segmenting image regions to finding a set of label assignment to tree nodes. We formulate the tree structure as a constrained conditional model to associate region merging with likelihoods predicted using an ensemble boundary classifier. Final segmentations can then be inferred by finding globally optimal solutions to the model efficiently. We also present an iterative training and testing algorithm that generates various tree structures and combines them to emphasize accurate boundaries by segmentation accumulation. Experiment results and comparisons with other recent methods on six public datasets demonstrate that our approach achieves the state-of-the-art region accuracy and is competitive in image segmentation without semantic priors.
T. Liu, M. Zhang, M. Javanmardi , N. Ramesh, T. Tasdizen.
“SSHMT: Semi-supervised Hierarchical Merge Tree for Electron Microscopy Image Segmentation,” In Lecture Notes in Computer Science, Vol. 9905, Springer International Publishing, pp. 144--159. 2016.
DOI: 10.1007/978-3-319-46448-0_9
Region-based methods have proven necessary for improving segmentation accuracy of neuronal structures in electron microscopy (EM) images. Most region-based segmentation methods use a scoring function to determine region merging. Such functions are usually learned with supervised algorithms that demand considerable ground truth data, which are costly to collect. We propose a semi-supervised approach that reduces this demand. Based on a merge tree structure, we develop a differentiable unsupervised loss term that enforces consistent predictions from the learned function. We then propose a Bayesian model that combines the supervised and the unsupervised information for probabilistic learning. The experimental results on three EM data sets demonstrate that by using a subset of only 3% to 7% of the entire ground truth data, our approach consistently performs close to the state-of-the-art supervised method with the full labeled data set, and significantly outperforms the supervised method with the same labeled subset.
F. Mesadi, M. Cetin, T. Tasdizen.
“Disjunctive normal level set: An efficient parametric implicit method,” In 2016 IEEE International Conference on Image Processing (ICIP), IEEE, September, 2016.
DOI: 10.1109/icip.2016.7533171
Level set methods are widely used for image segmentation because of their capability to handle topological changes. In this paper, we propose a novel parametric level set method called Disjunctive Normal Level Set (DNLS), and apply it to both two phase (single object) and multiphase (multi-object) image segmentations. The DNLS is formed by union of polytopes which themselves are formed by intersections of half-spaces. The proposed level set framework has the following major advantages compared to other level set methods available in the literature. First, segmentation using DNLS converges much faster. Second, the DNLS level set function remains regular throughout its evolution. Third, the proposed multiphase version of the DNLS is less sensitive to initialization, and its computational cost and memory requirement remains almost constant as the number of objects to be simultaneously segmented grows. The experimental results show the potential of the proposed method.
M. Sajjadi, S.M. Seyedhosseini, T. Tasdizen.
“Disjunctive Normal Networks,” In Neurocomputing, Vol. 218, Elsevier BV, pp. 276--285. Dec, 2016.
DOI: 10.1016/j.neucom.2016.08.047
Artificial neural networks are powerful pattern classifiers. They form the basis of the highly successful and popular Convolutional Networks which offer the state-of-the-art performance on several computer visions tasks. However, in many general and non-vision tasks, neural networks are surpassed by methods such as support vector machines and random forests that are also easier to use and faster to train. One reason is that the backpropagation algorithm, which is used to train artificial neural networks, usually starts from a random weight initialization which complicates the optimization process leading to long training times and increases the risk of stopping in a poor local minima. Several initialization schemes and pre-training methods have been proposed to improve the efficiency and performance of training a neural network. However, this problem arises from the architecture of neural networks. We use the disjunctive normal form and approximate the boolean conjunction operations with products to construct a novel network architecture. The proposed model can be trained by minimizing an error function and it allows an effective and intuitive initialization which avoids poor local minima. We show that the proposed structure provides efficient coverage of the decision space which leads to state-of-the art classification accuracy and fast training times.
M. Sajjadi, M. Javanmardi, T. Tasdizen.
“Regularization With Stochastic Transformations and Perturbations for Deep Semi-Supervised Learning,” In CoRR, Vol. abs/1606.04586, 2016.
Effective convolutional neural networks are trained on large sets of labeled data. However, creating large labeled datasets is a very costly and time-consuming task. Semi-supervised learning uses unlabeled data to train a model with higher accuracy when there is a limited set of labeled data available. In this paper, we consider the problem of semi-supervised learning with convolutional neural networks. Techniques such as randomized data augmentation, dropout and random max-pooling provide better generalization and stability for classifiers that are trained using gradient descent. Multiple passes of an individual sample through the network might lead to different predictions due to the non-deterministic behavior of these techniques. We propose an unsupervised loss function that takes advantage of the stochastic nature of these methods and minimizes the difference between the predictions of multiple passes of a training sample through the network. We evaluate the proposed method on several benchmark datasets.
M. Sajjadi, M. Javanmardi, T. Tasdizen.
“Mutual exclusivity loss for semi-supervised deep learning,” In 2016 IEEE International Conference on Image Processing (ICIP), IEEE, September, 2016.
In this paper we consider the problem of semi-supervised learning with deep Convolutional Neural Networks (ConvNets). Semi-supervised learning is motivated on the observation that unlabeled data is cheap and can be used to improve the accuracy of classifiers. In this paper we propose an unsupervised regularization term that explicitly forces the classifier's prediction for multiple classes to be mutually-exclusive and effectively guides the decision boundary to lie on the low density space between the manifolds corresponding to different classes of data. Our proposed approach is general and can be used with any backpropagation-based learning method. We show through different experiments that our method can improve the object recognition performance of ConvNets using unlabeled data.
S.M. Seyedhosseini, T. Tasdizen.
“Semantic Image Segmentation with Contextual Hierarchical Models,” In IEEE Transactions on Pattern Analysis and Machine Intelligence, Vol. 38, No. 5, IEEE, pp. 951--964. May, 2016.
DOI: 10.1109/tpami.2015.2473846
Semantic segmentation is the problem of assigning an object label to each pixel. It unifies the image segmentation and object recognition problems. The importance of using contextual information in semantic segmentation frameworks has been widely realized in the field. We propose a contextual framework, called contextual hierarchical model (CHM), which learns contextual information in a hierarchical framework for semantic segmentation. At each level of the hierarchy, a classifier is trained based on downsampled input images and outputs of previous levels. Our model then incorporates the resulting multi-resolution contextual information into a classifier to segment the input image at original resolution. This training strategy allows for optimization of a joint posterior probability at multiple resolutions through the hierarchy. Contextual hierarchical model is purely based on the input image patches and does not make use of any fragments or shape examples. Hence, it is applicable to a variety of problems such as object segmentation and edge detection. We demonstrate that CHM performs at par with state-of-the-art on Stanford background and Weizmann horse datasets. It also outperforms state-of-the-art edge detection methods on NYU depth dataset and achieves state-of-the-art on Berkeley segmentation dataset (BSDS 500).
2015
I. Arganda-Carreras, S.C. Turaga, D.R. Berger, D. Ciresan, A. Giusti, L.M. Gambardella, J. Schmidhuber, D. Laptev, S. Dwivedi, J. Buhmann, T. Liu, M. Seyedhosseini, T. Tasdizen, L. Kamentsky, R. Burget, V. Uher, X. Tan, C. Sun, T.D. Pham, E. Bas, M.G. Uzunbas, A. Cardona, J. Schindelin, H.S. Seung.
“Crowdsourcing the creation of image segmentation algorithms for connectomics,” In Frontiers in Neuroanatomy, Vol. 9, Frontiers Media SA, Nov, 2015.
DOI: 10.3389/fnana.2015.00142
To stimulate progress in automating the reconstruction of neural circuits, we organized the first international challenge on 2D segmentation of electron microscopic (EM) images of the brain. Participants submitted boundary maps predicted for a test set of images, and were scored based on their agreement with a consensus of human expert annotations. The winning team had no prior experience with EM images, and employed a convolutional network. This "deep learning" approach has since become accepted as a standard for segmentation of EM images. The challenge has continued to accept submissions, and the best so far has resulted from cooperation between two teams. The challenge has probably saturated, as algorithms cannot progress beyond limits set by ambiguities inherent in 2D scoring and the size of the test dataset. Retrospective evaluation of the challenge scoring system reveals that it was not sufficiently robust to variations in the widths of neurite borders. We propose a solution to this problem, which should be useful for a future 3D segmentation challenge.
E. Erdil, A.O. Argunsah, T. Tasdizen, D. Unay, M. Cetin.
“A joint classification and segmentation approach for dendritic spine segmentation in 2-photon microscopy images,” In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), IEEE, April, 2015.
DOI: 10.1109/isbi.2015.7163992
Shape priors have been successfully used in challenging biomedical imaging problems. However when the shape distribution involves multiple shape classes, leading to a multimodal shape density, effective use of shape priors in segmentation becomes more challenging. In such scenarios, knowing the class of the shape can aid the segmentation process, which is of course unknown a priori. In this paper, we propose a joint classification and segmentation approach for dendritic spine segmentation which infers the class of the spine during segmentation and adapts the remaining segmentation process accordingly. We evaluate our proposed approach on 2-photon microscopy images containing dendritic spines and compare its performance quantitatively to an existing approach based on nonparametric shape priors. Both visual and quantitative results demonstrate the effectiveness of our approach in dendritic spine segmentation.
M.U. Ghani, S.D. Kanik, A.O. Argunsah, T. Tasdizen, D. Unay, M. Cetin.
“Dendritic spine shape classification from two-photon microscopy images,” In 2015 23nd Signal Processing and Communications Applications Conference (SIU), IEEE, May, 2015.
DOI: 10.1109/siu.2015.7129985
Functional properties of a neuron are coupled with its morphology, particularly the morphology of dendritic spines. Spine volume has been used as the primary morphological parameter in order the characterize the structure and function coupling. However, this reductionist approach neglects the rich shape repertoire of dendritic spines. First step to incorporate spine shape information into functional coupling is classifying main spine shapes that were proposed in the literature. Due to the lack of reliable and fully automatic tools to analyze the morphology of the spines, such analysis is often performed manually, which is a laborious and time intensive task and prone to subjectivity. In this paper we present an automated approach to extract features using basic image processing techniques, and classify spines into mushroom or stubby by applying machine learning algorithms. Out of 50 manually segmented mushroom and stubby spines, Support Vector Machine was able to classify 98% of the spines correctly.
C. Jones, T. Liu, N.W. Cohan, M. Ellisman, T. Tasdizen.
“Efficient semi-automatic 3D segmentation for neuron tracing in electron microscopy images,” In Journal of Neuroscience Methods, Vol. 246, Elsevier BV, pp. 13--21. May, 2015.
DOI: 10.1016/j.jneumeth.2015.03.005
Background
In the area of connectomics, there is a significant gap between the time required for data acquisition and dense reconstruction of the neural processes contained in the same dataset. Automatic methods are able to eliminate this timing gap, but the state-of-the-art accuracy so far is insufficient for use without user corrections. If completed naively, this process of correction can be tedious and time consuming.
New method
We present a new semi-automatic method that can be used to perform 3D segmentation of neurites in EM image stacks. It utilizes an automatic method that creates a hierarchical structure for recommended merges of superpixels. The user is then guided through each predicted region to quickly identify errors and establish correct links.
Results
We tested our method on three datasets with both novice and expert users. Accuracy and timing were compared with published automatic, semi-automatic, and manual results.
Comparison with existing methods
Post-automatic correction methods have also been used in Mishchenko et al. (2010) and Haehn et al. (2014). These methods do not provide navigation or suggestions in the manner we present. Other semi-automatic methods require user input prior to the automatic segmentation such as Jeong et al. (2009) and Cardona et al. (2010) and are inherently different than our method.
Conclusion
Using this method on the three datasets, novice users achieved accuracy exceeding state-of-the-art automatic results, and expert users achieved accuracy on par with full manual labeling but with a 70% time improvement when compared with other examples in publication.
F. Mesadi, M. Cetin, T. Tasdizen.
“Disjunctive Normal Shape and Appearance Priors with Applications to Image Segmentation,” In Lecture Notes in Computer Science, Springer International Publishing, pp. 703--710. 2015.
ISBN: 978-3-319-24574-4
DOI: 10.1007/978-3-319-24574-4_84
The use of appearance and shape priors in image segmentation is known to improve accuracy; however, existing techniques have several drawbacks. Active shape and appearance models require landmark points and assume unimodal shape and appearance distributions. Level set based shape priors are limited to global shape similarity. In this paper, we present a novel shape and appearance priors for image segmentation based on an implicit parametric shape representation called disjunctive normal shape model (DNSM). DNSM is formed by disjunction of conjunctions of half-spaces defined by discriminants. We learn shape and appearance statistics at varying spatial scales using nonparametric density estimation. Our method can generate a rich set of shape variations by locally combining training shapes. Additionally, by studying the intensity and texture statistics around each discriminant of our shape model, we construct a local appearance probability map. Experiments carried out on both medical and natural image datasets show the potential of the proposed method.
N. Ramesh, F. Mesadi, M. Cetin, T. Tasdizen.
“Disjunctive normal shape models,” In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), IEEE, April, 2015.
DOI: 10.1109/isbi.2015.7164170
A novel implicit parametric shape model is proposed for segmentation and analysis of medical images. Functions representing the shape of an object can be approximated as a union of N polytopes. Each polytope is obtained by the intersection of M half-spaces. The shape function can be approximated as a disjunction of conjunctions, using the disjunctive normal form. The shape model is initialized using seed points defined by the user. We define a cost function based on the Chan-Vese energy functional. The model is differentiable, hence, gradient based optimization algorithms are used to find the model parameters.
M. Sajjadi, M. Seyedhosseini,, T. Tasdizen.
“Nonlinear Regression with Logistic Product Basis Networks,” In IEEE Signal Processing Letters, Vol. 22, No. 8, IEEE, pp. 1011--1015. Aug, 2015.
DOI: 10.1109/lsp.2014.2380791
We introduce a novel general regression model that is based on a linear combination of a new set of non-local basis functions that forms an effective feature space. We propose a training algorithm that learns all the model parameters simultaneously and offer an initialization scheme for parameters of the basis functions. We show through several experiments that the proposed method offers better coverage for high-dimensional space compared to local Gaussian basis functions and provides competitive performance in comparison to other state-of-the-art regression methods.
M. Seyedhosseini , T. Tasdizen.
“Disjunctive normal random forests,” In Pattern Recognition, Vol. 48, No. 3, Elsevier BV, pp. 976--983. March, 2015.
DOI: 10.1016/j.patcog.2014.08.023
We develop a novel supervised learning/classification method, called disjunctive normal random forest (DNRF). A DNRF is an ensemble of randomly trained disjunctive normal decision trees (DNDT). To construct a DNDT, we formulate each decision tree in the random forest as a disjunction of rules, which are conjunctions of Boolean functions. We then approximate this disjunction of conjunctions with a differentiable function and approach the learning process as a risk minimization problem that incorporates the classification error into a single global objective function. The minimization problem is solved using gradient descent. DNRFs are able to learn complex decision boundaries and achieve low generalization error. We present experimental results demonstrating the improved performance of DNDTs and DNRFs over conventional decision trees and random forests. We also show the superior performance of DNRFs over state-of-the-art classification methods on benchmark datasets.
S.M. Seyedhosseini, S. Shushruth, T. Davis, J.M. Ichida, P.A. House, B. Greger, A. Angelucci, T. Tasdizen.
“Informative features of local field potential signals in primary visual cortex during natural image stimulation,” In Journal of Neurophysiology, Vol. 113, No. 5, American Physiological Society, pp. 1520--1532. March, 2015.
DOI: 10.1152/jn.00278.2014
The local field potential (LFP) is of growing importance in neurophysiology as a metric of network activity and as a readout signal for use in brain-machine interfaces. However, there are uncertainties regarding the kind and visual field extent of information carried by LFP signals, as well as the specific features of the LFP signal conveying such information, especially under naturalistic conditions. To address these questions, we recorded LFP responses to natural images in V1 of awake and anesthetized macaques using Utah multielectrode arrays. First, we have shown that it is possible to identify presented natural images from the LFP responses they evoke using trained Gabor wavelet (GW) models. Because GW models were devised to explain the spiking responses of V1 cells, this finding suggests that local spiking activity and LFPs (thought to reflect primarily local synaptic activity) carry similar visual information. Second, models trained on scalar metrics, such as the evoked LFP response range, provide robust image identification, supporting the informative nature of even simple LFP features. Third, image identification is robust only for the first 300 ms following image presentation, and image information is not restricted to any of the spectral bands. This suggests that the short-latency broadband LFP response carries most information during natural scene viewing. Finally, best image identification was achieved by GW models incorporating information at the scale of ∼0.5° in size and trained using four different orientations. This suggests that during natural image viewing, LFPs carry stimulus-specific information at spatial scales corresponding to few orientation columns in macaque V1.
2014
T. Liu, C. Jones, M. Seyedhosseini, T. Tasdizen.
“A modular hierarchical approach to 3D electron microscopy image segmentation,” In Journal of Neuroscience Methods, Vol. 226, No. 15, pp. 88--102. April, 2014.
DOI: 10.1016/j.jneumeth.2014.01.022
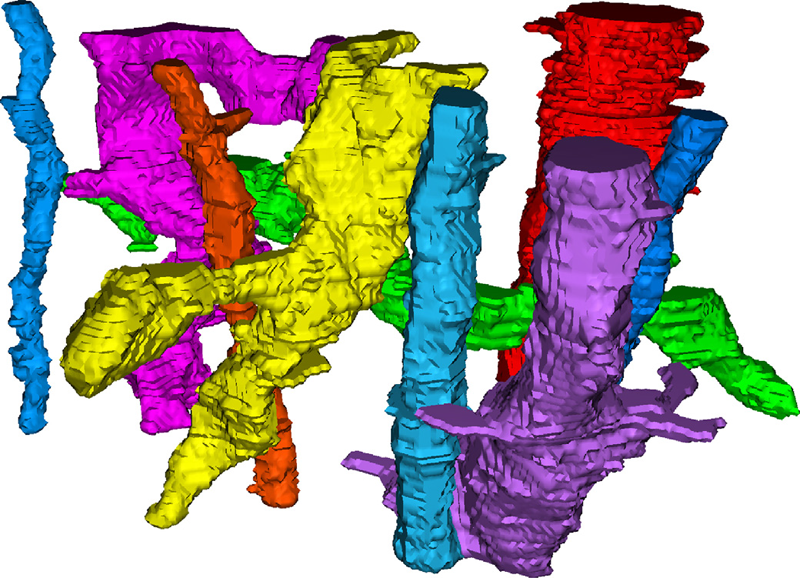
Keywords: Image segmentation, Electron microscopy, Hierarchical segmentation, Semi-automatic segmentation, Neuron reconstruction
A. J. Perez, M. Seyedhosseini, T. J. Deerinck, E. A. Bushong, S. Panda, T. Tasdizen, M. H. Ellisman.
“A workflow for the automatic segmentation of organelles in electron microscopy image stacks,” In Frontiers in Neuroanatomy, Vol. 8, No. 126, 2014.
DOI: 10.3389/fnana.2014.00126
Electron microscopy (EM) facilitates analysis of the form, distribution, and functional status of key organelle systems in various pathological processes, including those associated with neurodegenerative disease. Such EM data often provide important new insights into the underlying disease mechanisms. The development of more accurate and efficient methods to quantify changes in subcellular microanatomy has already proven key to understanding the pathogenesis of Parkinson's and Alzheimer's diseases, as well as glaucoma. While our ability to acquire large volumes of 3D EM data is progressing rapidly, more advanced analysis tools are needed to assist in measuring precise three-dimensional morphologies of organelles within data sets that can include hundreds to thousands of whole cells. Although new imaging instrument throughputs can exceed teravoxels of data per day, image segmentation and analysis remain significant bottlenecks to achieving quantitative descriptions of whole cell structural organellomes. Here, we present a novel method for the automatic segmentation of organelles in 3D EM image stacks. Segmentations are generated using only 2D image information, making the method suitable for anisotropic imaging techniques such as serial block-face scanning electron microscopy (SBEM). Additionally, no assumptions about 3D organelle morphology are made, ensuring the method can be easily expanded to any number of structurally and functionally diverse organelles. Following the presentation of our algorithm, we validate its performance by assessing the segmentation accuracy of different organelle targets in an example SBEM dataset and demonstrate that it can be efficiently parallelized on supercomputing resources, resulting in a dramatic reduction in runtime.
A. Perez, M. Seyedhosseini, T. Tasdizen, M. Ellisman.
“Automated workflows for the morphological characterization of organelles in electron microscopy image stacks (LB72),” In The FASEB Journal, Vol. 28, No. 1 Supplement LB72, April, 2014.
Advances in three-dimensional electron microscopy (EM) have facilitated the collection of image stacks with a field-of-view that is large enough to cover a significant percentage of anatomical subdivisions at nano-resolution. When coupled with enhanced staining protocols, such techniques produce data that can be mined to establish the morphologies of all organelles across hundreds of whole cells in their in situ environments. Although instrument throughputs are approaching terabytes of data per day, image segmentation and analysis remain significant bottlenecks in achieving quantitative descriptions of whole cell organellomes. Here we describe computational workflows that achieve the automatic segmentation of organelles from regions of the central nervous system by applying supervised machine learning algorithms to slices of serial block-face scanning EM (SBEM) datasets. We also demonstrate that our workflows can be parallelized on supercomputing resources, resulting in a dramatic reduction of their run times. These methods significantly expedite the development of anatomical models at the subcellular scale and facilitate the study of how these models may be perturbed following pathological insults.
Page 4 of 9
