SCI Publications
2014
N. Ramesh, T. Tasdizen.
“Cell tracking using particle filters with implicit convex shape model in 4D confocal microscopy images,” In 2014 IEEE International Conference on Image Processing (ICIP), IEEE, Oct, 2014.
DOI: 10.1109/icip.2014.7025089
Bayesian frameworks are commonly used in tracking algorithms. An important example is the particle filter, where a stochastic motion model describes the evolution of the state, and the observation model relates the noisy measurements to the state. Particle filters have been used to track the lineage of cells. Propagating the shape model of the cell through the particle filter is beneficial for tracking. We approximate arbitrary shapes of cells with a novel implicit convex function. The importance sampling step of the particle filter is defined using the cost associated with fitting our implicit convex shape model to the observations. Our technique is capable of tracking the lineage of cells for nonmitotic stages. We validate our algorithm by tracking the lineage of retinal and lens cells in zebrafish embryos.
M. Seyedhosseini, T. Tasdizen.
“Disjunctive normal random forests,” In Pattern Recognition, September, 2014.
DOI: 10.1016/j.patcog.2014.08.023
We develop a novel supervised learning/classification method, called disjunctive normal random forest (DNRF). A DNRF is an ensemble of randomly trained disjunctive normal decision trees (DNDT). To construct a DNDT, we formulate each decision tree in the random forest as a disjunction of rules, which are conjunctions of Boolean functions. We then approximate this disjunction of conjunctions with a differentiable function and approach the learning process as a risk minimization problem that incorporates the classification error into a single global objective function. The minimization problem is solved using gradient descent. DNRFs are able to learn complex decision boundaries and achieve low generalization error. We present experimental results demonstrating the improved performance of DNDTs and DNRFs over conventional decision trees and random forests. We also show the superior performance of DNRFs over state-of-the-art classification methods on benchmark datasets.
Keywords: Random forest, Decision tree, Classifier, Supervised learning, Disjunctive normal form
M. Seyedhosseini, T. Tasdizen.
“Scene Labeling with Contextual Hierarchical Models,” In CoRR, Vol. abs/1402.0595, 2014.
T. Tasdizen, M. Seyedhosseini, T. Liu, C. Jones, E. Jurrus.
“Image Segmentation for Connectomics Using Machine Learning,” In Computational Intelligence in Biomedical Imaging, Edited by Suzuki, Kenji, Springer New York, pp. 237--278. 2014.
ISBN: 978-1-4614-7244-5
DOI: 10.1007/978-1-4614-7245-2_10
Reconstruction of neural circuits at the microscopic scale of individual neurons and synapses, also known as connectomics, is an important challenge for neuroscience. While an important motivation of connectomics is providing anatomical ground truth for neural circuit models, the ability to decipher neural wiring maps at the individual cell level is also important in studies of many neurodegenerative diseases. Reconstruction of a neural circuit at the individual neuron level requires the use of electron microscopy images due to their extremely high resolution. Computational challenges include pixel-by-pixel annotation of these images into classes such as cell membrane, mitochondria and synaptic vesicles and the segmentation of individual neurons. State-of-the-art image analysis solutions are still far from the accuracy and robustness of human vision and biologists are still limited to studying small neural circuits using mostly manual analysis. In this chapter, we describe our image analysis pipeline that makes use of novel supervised machine learning techniques to tackle this problem.
2013
C. Jones, T. Liu, M. Ellisman, T. Tasdizen.
“Semi-Automatic Neuron Segmentation in Electron Microscopy Images Via Sparse Labeling,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 1304--1307. April, 2013.
DOI: 10.1109/ISBI.2013.6556771
C. Jones, M. Seyedhosseini, M. Ellisman, T. Tasdizen.
“Neuron Segmentation in Electron Microscopy Images Using Partial Differential Equations,” In Proceedings of 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 1457--1460. April, 2013.
DOI: 10.1109/ISBI.2013.6556809
T. Liu, M. Seyedhosseini, M. Ellisman, T. Tasdizen.
“Watershed Merge Forest Classification for Electron Microscopy Image Stack Segmentation,” In Proceedings of the 2013 International Conference on Image Processing, 2013.
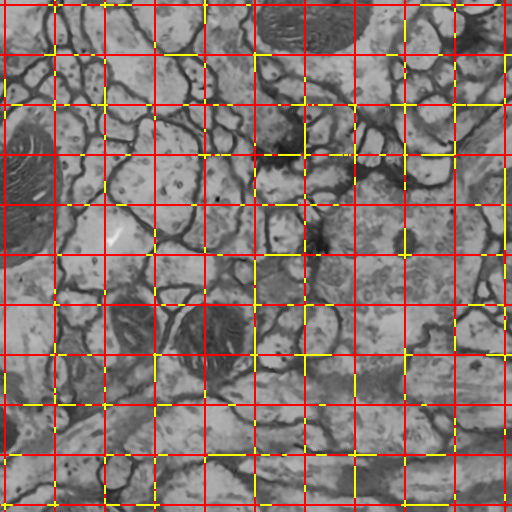
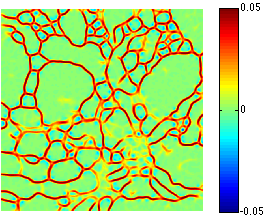
Automated electron microscopy (EM) image analysis techniques can be tremendously helpful for connectomics research. In this paper, we extend our previous work [1] and propose a fully automatic method to utilize inter-section information for intra-section neuron segmentation of EM image stacks. A watershed merge forest is built via the watershed transform with each tree representing the region merging hierarchy of one 2D section in the stack. A section classifier is learned to identify the most likely region correspondence between adjacent sections. The inter-section information from such correspondence is incorporated to update the potentials of tree nodes. We resolve the merge forest using these potentials together with consistency constraints to acquire the final segmentation of the whole stack. We demonstrate that our method leads to notable segmentation accuracy improvement by experimenting with two types of EM image data sets.
N. Ramesh, T. Tasdizen.
“Three-dimensional alignment and merging of confocal microscopy stacks,” In 2013 IEEE International Conference on Image Processing, IEEE, September, 2013.
DOI: 10.1109/icip.2013.6738297
We describe an efficient, robust, automated method for image alignment and merging of translated, rotated and flipped con-focal microscopy stacks. The samples are captured in both directions (top and bottom) to increase the SNR of the individual slices. We identify the overlapping region of the two stacks by using a variable depth Maximum Intensity Projection (MIP) in the z dimension. For each depth tested, the MIP images gives an estimate of the angle of rotation between the stacks and the shifts in the x and y directions using the Fourier Shift property in 2D. We use the estimated rotation angle, shifts in the x and y direction and align the images in the z direction. A linear blending technique based on a sigmoidal function is used to maximize the information from the stacks and combine them. We get maximum information gain as we combine stacks obtained from both directions.
M. Seyedhosseini, M. Ellisman, T. Tasdizen.
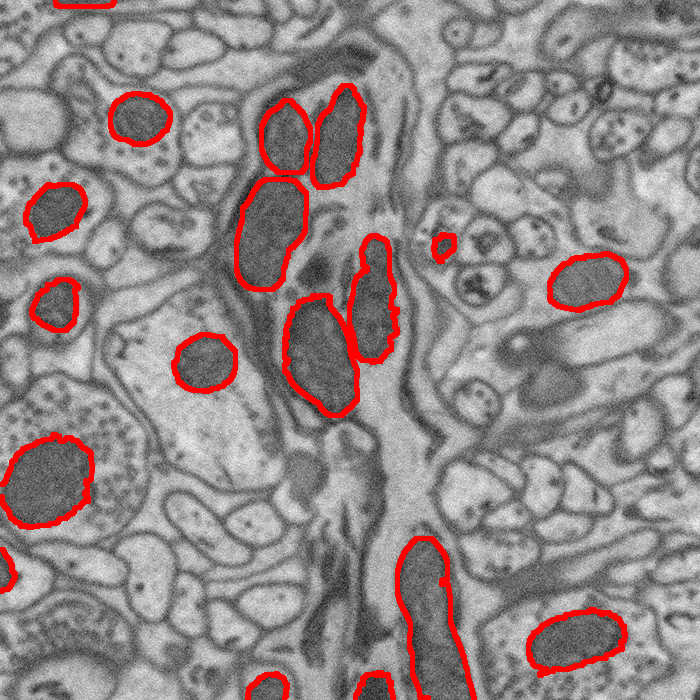
“Segmentation of Mitochondria in Electron Microscopy Images using Algebraic Curves,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 860--863. 2013.
DOI: 10.1109/ISBI.2013.6556611
M. Seyedhosseini, T. Tasdizen.
“Multi-class Multi-scale Series Contextual Model for Image Segmentation,” In IEEE Transactions on Image Processing, Vol. PP, No. 99, 2013.
DOI: 10.1109/TIP.2013.2274388
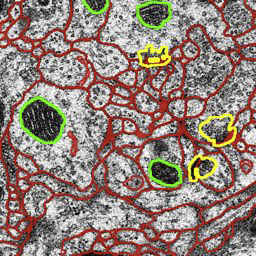
M. Seyedhosseini, M. Sajjadi, T. Tasdizen.
“Image Segmentation with Cascaded Hierarchical Models and Logistic Disjunctive Normal Networks,” In Proceedings of the IEEE International Conference on Computer Vison (ICCV 2013), pp. (accepted). 2013.
Contextual information plays an important role in solving vision problems such as image segmentation. However, extracting contextual information and using it in an effective way remains a difficult problem. To address this challenge, we propose a multi-resolution contextual framework, called cascaded hierarchical model (CHM), which learns contextual information in a hierarchical framework for image segmentation. At each level of the hierarchy, a classifier is trained based on downsampled input images and outputs of previous levels. Our model then incorporates the resulting multi-resolution contextual information into a classifier to segment the input image at original resolution. We repeat this procedure by cascading the hierarchical framework to improve the segmentation accuracy. Multiple classifiers are learned in the CHM; therefore, a fast and accurate classifier is required to make the training tractable. The classifier also needs to be robust against overfitting due to the large number of parameters learned during training. We introduce a novel classification scheme, called logistic disjunctive normal networks (LDNN), which consists of one adaptive layer of feature detectors implemented by logistic sigmoid functions followed by two fixed layers of logical units that compute conjunctions and disjunctions, respectively. We demonstrate that LDNN outperforms state-of-theart classifiers and can be used in the CHM to improve object segmentation performance.
2012
E.V.R. DiBella, T. Tasdizen.
“Edge enhanced spatio-temporal constrained reconstruction of undersampled dynamic contrast enhanced radial MRI,” In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 704--707. 2012.
DOI: 10.1109/ISBI.2010.5490077
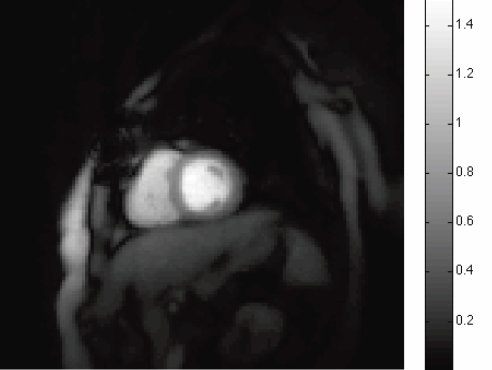
L. Hogrebe, A.R.C. Paiva, E. Jurrus, C. Christensen, M. Bridge, L. Dai, R.L. Pfeiffer, P.R. Hof, B. Roysam, J.R. Korenberg, T. Tasdizen.
“Serial section registration of axonal confocal microscopy datasets for long-range neural circuit reconstruction,” In Journal of Neuroscience Methods, Vol. 207, No. 2, pp. 200--210. 2012.
DOI: 10.1016/j.jneumeth.2012.03.002
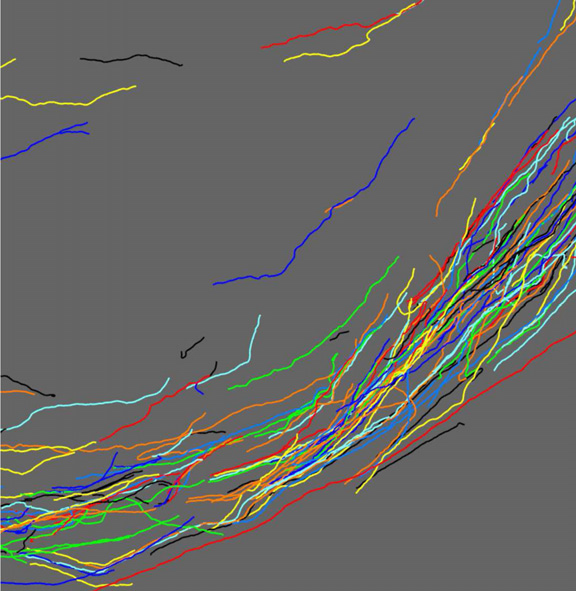
S.K. Iyer, T. Tasdizen, E.V.R. DiBella.
“Edge Enhanced Spatio-Temporal Constrained Reconstruction of Undersampled Dynamic Contrast Enhanced Radial MRI,” In Magnetic Resonance Imaging, Vol. 30, No. 5, pp. 610--619. 2012.
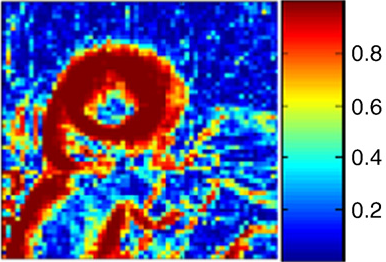
Keywords: MRI, Reconstruction, Edge enhanced, Compressed sensing, Regularization, Cardiac perfusion
E. Jurrus, S. Watanabe, R.J. Giuly, A.R.C. Paiva, M.H. Ellisman, E.M. Jorgensen, T. Tasdizen.
“Semi-Automated Neuron Boundary Detection and Nonbranching Process Segmentation in Electron Microscopy Images,” In Neuroinformatics, pp. (published online). 2012.
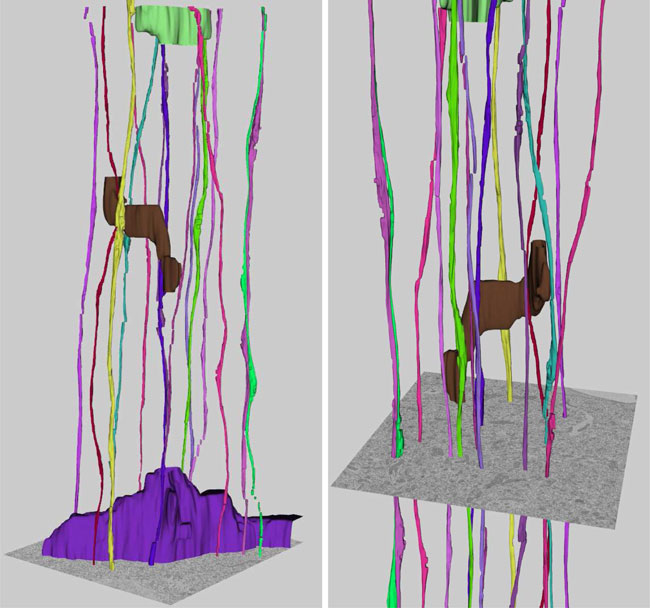
T. Liu, E. Jurrus, M. Seyedhosseini, M. Ellisman, T. Tasdizen.
“Watershed Merge Tree Classification for Electron Microscopy Image Segmentation,” In Proceedings of the 21st International Conference on Pattern Recognition (ICPR), pp. 133--137. 2012.
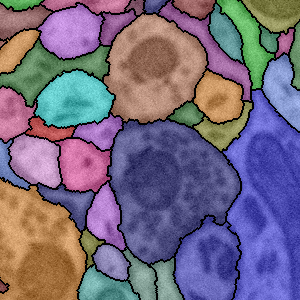
A.R.C. Paiva, T. Tasdizen.
“Fingerprint Image Segmentation using Data Manifold Characteristic Features,” In International Journal of Pattern Recognition and Artificial Intelligence, Vol. 26, No. 4, pp. (23 pages). 2012.
DOI: 10.1142/S0218001412560101

Keywords: Fingerprint segmentation, manifold characterization, feature extraction, dimensionality reduction
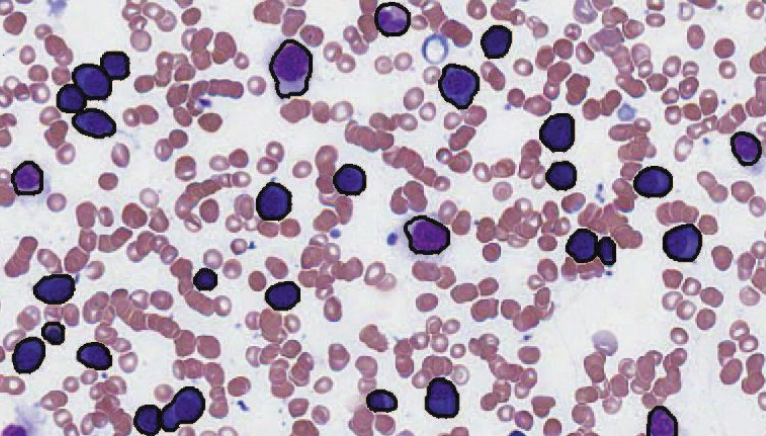
N. Ramesh, B. J. Dangott, M. Salama, T. Tasdizen.
“Segmentation and Two-Step Classification of White Blood Cells in Peripheral Blood Smear,” In Journal of Pathology Informatics, Vol. 3, No. 13, 2012.
An automated system for differential white blood cell (WBC) counting based on morphology can make manual differential leukocyte counts faster and less tedious for pathologists and laboratory professionals. We present an automated system for isolation and classification of WBCs in manually prepared, Wright stained, peripheral blood smears from whole slide images (WSI). Methods: A simple, classification scheme using color information and morphology is proposed. The performance of the algorithm was evaluated by comparing our proposed method with a hematopathologist's visual classification. The isolation algorithm was applied to 1938 subimages of WBCs, 1804 of them were accurately isolated. Then, as the first step of a two-step classification process, WBCs were broadly classified into cells with segmented nuclei and cells with nonsegmented nuclei. The nucleus shape is one of the key factors in deciding how to classify WBCs. Ambiguities associated with connected nuclear lobes are resolved by detecting maximum curvature points and partitioning them using geometric rules. The second step is to define a set of features using the information from the cytoplasm and nuclear regions to classify WBCs using linear discriminant analysis. This two-step classification approach stratifies normal WBC types accurately from a whole slide image. Results: System evaluation is performed using a 10-fold cross-validation technique. Confusion matrix of the classifier is presented to evaluate the accuracy for each type of WBC detection. Experiments show that the two-step classification implemented achieves a 93.9\% overall accuracy in the five subtype classification. Conclusion: Our methodology achieves a semiautomated system for the detection and classification of normal WBCs from scanned WSI. Further studies will be focused on detecting and segmenting abnormal WBCs, comparison of 20x and 40x data, and expanding the applications for bone marrow aspirates.
N. Ramesh, M.E. Salama, T. Tasdizen.
“Segmentation of Haematopoeitic Cells in Bone Marrow Using Circle Detection and Splitting Techniques,” In 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 206--209. 2012.
DOI: 10.1109/ISBI.2012.6235520
2011
J.R. Anderson, B.W. Jones, C.B. Watt, M.V. Shaw, J.-H. Yang, D. DeMill, J.S. Lauritzen, Y. Lin, K.D. Rapp, D. Mastronarde, P. Koshevoy, B. Grimm, T. Tasdizen, R.T. Whitaker, R.E. Marc.
“Exploring the Retinal Connectome,” In Molecular Vision, Vol. 17, pp. 355--379. 2011.
PubMed ID: 21311605
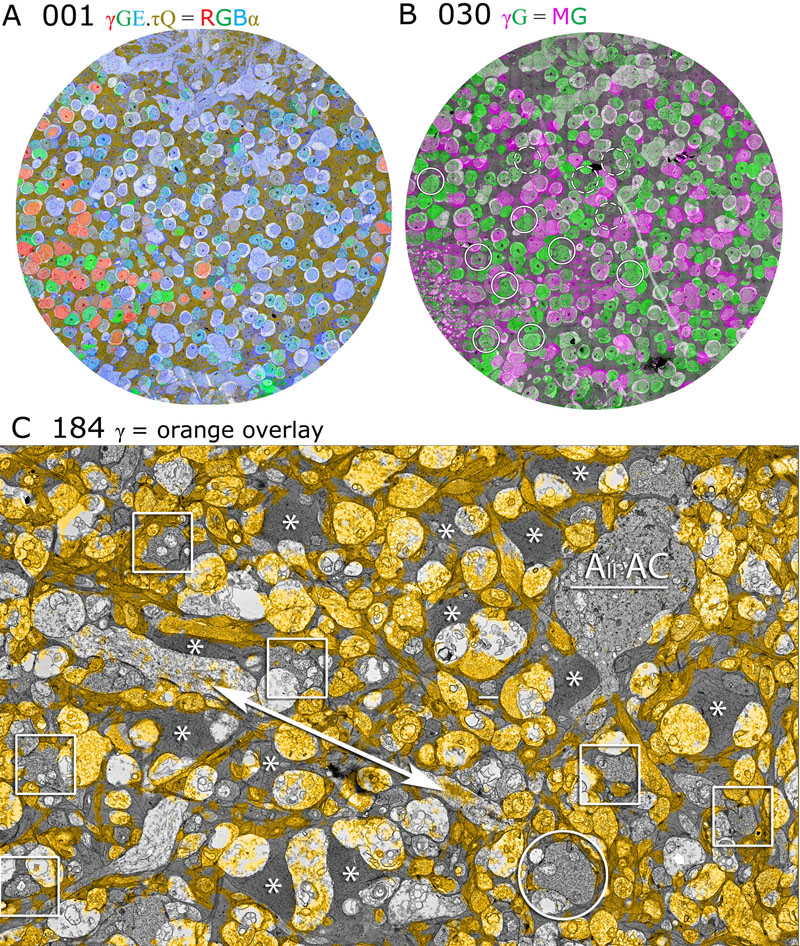
Purpose: A connectome is a comprehensive description of synaptic connectivity for a neural domain. Our goal was to produce a connectome data set for the inner plexiform layer of the mammalian retina. This paper describes our first retinal connectome, validates the method, and provides key initial findings.
Methods: We acquired and assembled a 16.5 terabyte connectome data set RC1 for the rabbit retina at .2 nm resolution using automated transmission electron microscope imaging, automated mosaicking, and automated volume registration. RC1 represents a column of tissue 0.25 mm in diameter, spanning the inner nuclear, inner plexiform, and ganglion cell layers. To enhance ultrastructural tracing, we included molecular markers for 4-aminobutyrate (GABA), glutamate, glycine, taurine, glutamine, and the in vivo activity marker, 1-amino-4-guanidobutane. This enabled us to distinguish GABAergic and glycinergic amacrine cells; to identify ON bipolar cells coupled to glycinergic cells; and to discriminate different kinds of bipolar, amacrine, and ganglion cells based on their molecular signatures and activity. The data set was explored and annotated with Viking, our multiuser navigation tool. Annotations were exported to additional applications to render cells, visualize network graphs, and query the database.
Results: Exploration of RC1 showed that the 2 nm resolution readily recapitulated well known connections and revealed several new features of retinal organization: (1) The well known AII amacrine cell pathway displayed more complexity than previously reported, with no less than 17 distinct signaling modes, including ribbon synapse inputs from OFF bipolar cells, wide-field ON cone bipolar cells and rod bipolar cells, and extensive input from cone-pathway amacrine cells. (2) The axons of most cone bipolar cells formed a distinct signal integration compartment, with ON cone bipolar cell axonal synapses targeting diverse cell types. Both ON and OFF bipolar cells receive axonal veto synapses. (3) Chains of conventional synapses were very common, with intercalated glycinergic-GABAergic chains and very long chains associated with starburst amacrine cells. Glycinergic amacrine cells clearly play a major role in ON-OFF crossover inhibition. (4) Molecular and excitation mapping clearly segregates ultrastructurally defined bipolar cell groups into different response clusters. (5) Finally, low-resolution electron or optical imaging cannot reliably map synaptic connections by process geometry, as adjacency without synaptic contact is abundant in the retina. Only direct visualization of synapses and gap junctions suffices.
Conclusions: Connectome assembly and analysis using conventional transmission electron microscopy is now practical for network discovery. Our surveys of volume RC1 demonstrate that previously studied systems such as the AII amacrine cell network involve more network motifs than previously known. The AII network, primarily considered a scotopic pathway, clearly derives ribbon synapse input from photopic ON and OFF cone bipolar cell networks and extensive photopic GABAergic amacrine cell inputs. Further, bipolar cells show extensive inputs and outputs along their axons, similar to multistratified nonmammalian bipolar cells. Physiologic evidence of significant ON-OFF channel crossover is strongly supported by our anatomic data, showing alternating glycine-to-GABA paths. Long chains of amacrine cell networks likely arise from homocellular GABAergic synapses between starburst amacrine cells. Deeper analysis of RC1 offers the opportunity for more complete descriptions of specific networks.
Keywords: neuroscience, retina, vision, blindness, visus, crcns
Page 5 of 9
