SCI Publications
2012
Q. Meng, J. Hall, H. Rutigliano, X. Zhou, B.R. Sessions, R. Stott, K. Panter, C.J. Davies, R. Ranjan, D. Dosdall, R.S. MacLeod, N. Marrouche, K.L. White, Z. Wang, I.A. Polejaeva.
“30 Generation of Cloned Transgenic Goats with Cardiac Specific Overexpression of Transforming Growth Factor β1,” In Reproduction, Fertility and Development, Vol. 25, No. 1, pp. 162--163. 2012.
DOI: 10.1071/RDv25n1Ab30
Transforming growth factor β1 (TGF-β1) has a potent profibrotic function and is central to signaling cascades involved in interstitial fibrosis, which plays a critical role in the pathobiology of cardiomyopathy and contributes to diastolic and systolic dysfunction. In addition, fibrotic remodeling is responsible for generation of re-entry circuits that promote arrhythmias (Bujak and Frangogiannis 2007 Cardiovasc. Res. 74, 184–195). Due to the small size of the heart, functional electrophysiology of transgenic mice is problematic. Large transgenic animal models have the potential to offer insights into conduction heterogeneity associated with fibrosis and the role of fibrosis in cardiovascular diseases. The goal of this study was to generate transgenic goats overexpressing an active form of TGFβ-1 under control of the cardiac-specific α-myosin heavy chain promoter (α-MHC). A pcDNA3.1DV5-MHC-TGF-β1cys33ser vector was constructed by subcloning the MHC-TGF-β1 fragment from the plasmid pUC-BM20-MHC-TGF-β1 (Nakajima et al. 2000 Circ. Res. 86, 571–579) into the pcDNA3.1D V5 vector. The Neon transfection system was used to electroporate primary goat fetal fibroblasts. After G418 selection and PCR screening, transgenic cells were used for SCNT. Oocytes were collected by slicing ovaries from an abattoir and matured in vitro in an incubator with 5\% CO2 in air. Cumulus cells were removed at 21 to 23 h post-maturation. Oocytes were enucleated by aspirating the first polar body and nearby cytoplasm by micromanipulation in Hepes-buffered SOF medium with 10 µg of cytochalasin B mL–1. Transgenic somatic cells were individually inserted into the perivitelline space and fused with enucleated oocytes using double electrical pulses of 1.8 kV cm–1 (40 µs each). Reconstructed embryos were activated by ionomycin (5 min) and DMAP and cycloheximide (CHX) treatments. Cloned embryos were cultured in G1 medium for 12 to 60 h in vitro and then transferred into synchronized recipient females. Pregnancy was examined by ultrasonography on day 30 post-transfer. A total of 246 cloned embryos were transferred into 14 recipients that resulted in production of 7 kids. The pregnancy rate was higher in the group cultured for 12 h compared with those cultured 36 to 60 h [44.4\% (n = 9) v. 20\% (n = 5)]. The kidding rates per embryo transferred of these 2 groups were 3.8\% (n = 156) and 1.1\% (n = 90), respectively. The PCR results confirmed that all the clones were transgenic. Phenotype characterization [e.g. gene expression, electrocardiogram (ECG), and magnetic resonance imaging (MRI)] is underway. We demonstrated successful production of transgenic goat via SCNT. To our knowledge, this is the first transgenic goat model produced for cardiovascular research.
M.D. Meyer, M. Sedlmair, T. Munzner.
“The Four-Level Nested Model Revisited: Blocks and Guidelines,” In Workshop on BEyond time and errors: novel evaLuation methods for Information Visualization (BELIV), IEEE VisWeek 2012, 2012.
P. Muralidharan, P.T. Fletcher.
“Sasaki Metrics for Analysis of Longitudinal Data on Manifolds,” In Proceedings of the 2012 IEEE conference on Computer Vision and Pattern Recognition (CVPR), pp. 1027--1034. 2012.
DOI: 10.1109/CVPR.2012.6247780
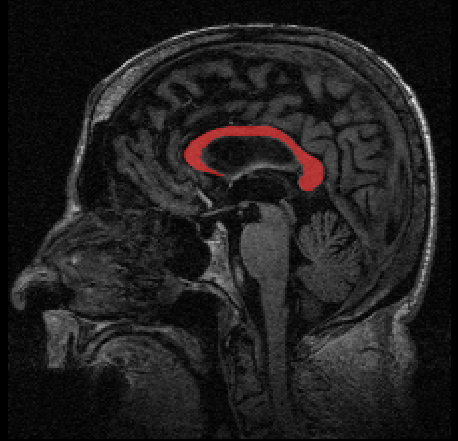
Longitudinal data arises in many applications in which the goal is to understand changes in individual entities over time. In this paper, we present a method for analyzing longitudinal data that take values in a Riemannian manifold. A driving application is to characterize anatomical shape changes and to distinguish between trends in anatomy that are healthy versus those that are due to disease. We present a generative hierarchical model in which each individual is modeled by a geodesic trend, which in turn is considered as a perturbation of the mean geodesic trend for the population. Each geodesic in the model can be uniquely parameterized by a starting point and velocity, i.e., a point in the tangent bundle. Comparison between these parameters is achieved through the Sasaki metric, which provides a natural distance metric on the tangent bundle. We develop a statistical hypothesis test for differences between two groups of longitudinal data by generalizing the Hotelling T2 statistic to manifolds. We demonstrate the ability of these methods to distinguish differences in shape changes in a comparison of longitudinal corpus callosum data in subjects with dementia versus healthily aging controls.
A. Narayan, D. Xiu.
“Stochastic Collocation Methods on Unstructured Grids in High Dimensions via Interpolation,” In SIAM Journal on Scientific Computing, Vol. 34, No. 3, pp. A1729–-A1752. 2012.
DOI: 10.1137/110854059
Keywords: stochastic collocation, polynomial chaos, interpolation, orthogonal polynomials
A. Narayan, Y. Marzouk, D. Xiu.
“Sequential Data Assimilation with Multiple Models,” In Journal of Computational Physics, Vol. 231, No. 19, pp. 6401--6418. 2012.
DOI: 10.1016/j.jcp.2012.06.002
Data assimilation is an essential tool for predicting the behavior of real physical systems given approximate simulation models and limited observations. For many complex systems, there may exist several models, each with different properties and predictive capabilities. It is desirable to incorporate multiple models into the assimilation procedure in order to obtain a more accurate prediction of the physics than any model alone can provide. In this paper, we propose a framework for conducting sequential data assimilation with multiple models and sources of data. The assimilated solution is a linear combination of all model predictions and data. One notable feature is that the combination takes the most general form with matrix weights. By doing so the method can readily utilize different weights in different sections of the solution state vectors, allow the models and data to have different dimensions, and deal with the case of a singular state covariance. We prove that the proposed assimilation method, termed direct assimilation, minimizes a variational functional, a generalized version of the one used in the classical Kalman filter. We also propose an efficient iterative assimilation method that assimilates two models at a time until all models and data are assimilated. The mathematical equivalence of the iterative method and the direct method is established. Numerical examples are presented to demonstrate the effectiveness of the new method.
Keywords: Uncertainty quantification, Data assimilation, Kalman filter, Model averaging
B. Nelson, E. Liu, R.M. Kirby, R. Haimes.
“ElVis: A System for the Accurate and Interactive Visualization of High-Order Finite Element Solutions,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 18, No. 12, pp. 2325--2334. Dec, 2012.
DOI: 10.1109/TVCG.2012.218
A.R.C. Paiva, T. Tasdizen.
“Fingerprint Image Segmentation using Data Manifold Characteristic Features,” In International Journal of Pattern Recognition and Artificial Intelligence, Vol. 26, No. 4, pp. (23 pages). 2012.
DOI: 10.1142/S0218001412560101

Keywords: Fingerprint segmentation, manifold characterization, feature extraction, dimensionality reduction
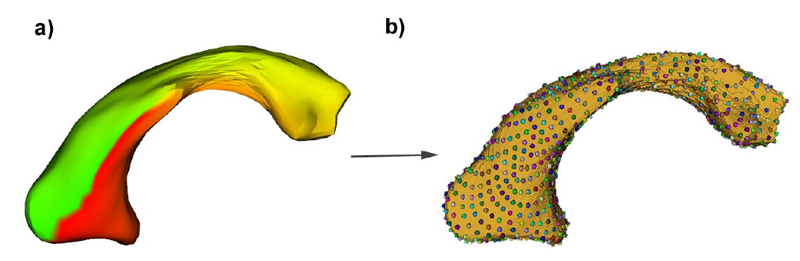
B. Paniagua, L. Bompard, J. Cates, R.T. Whitaker, M. Datar, C. Vachet, M. Styner.
“Combined SPHARM-PDM and entropy-based particle systems shape analysis framework,” In Medical Imaging 2012: Biomedical Applications in Molecular, Structural, and Functional Imaging, SPIE Intl Soc Optical Eng, March, 2012.
DOI: 10.1117/12.911228
PubMed ID: 24027625
PubMed Central ID: PMC3766973
C Partl, A Lex, M Streit, D Kalkofen, K Kashofer, D Schmalstieg.
“enRoute: Dynamic Path Extraction from Biological Pathway Maps for In-Depth Experimental Data Analysis,” In Proceedings of the IEEE Symposium on Biological Data Visualization (BioVis '12), IEEE, pp. 107--114. 2012.
DOI: 10.1109/BioVis.2012.6378600
Pathway maps are an important source of information when analyzing functional implications of experimental data on biological processes. However, associating large quantities of data with nodes on a pathway map and allowing in depth-analysis at the same time is a challenging task. While a wide variety of approaches for doing so exist, they either do not scale beyond a few experiments or fail to represent the pathway appropriately. To remedy this, we introduce enRoute, a new approach for interactively exploring experimental data along paths that are dynamically extracted from pathways. By showing an extracted path side-by-side with experimental data, enRoute can present large amounts of data for every pathway node. It can visualize hundreds of samples, dozens of experimental conditions, and even multiple datasets capturing different aspects of a node at the same time. Another important property of this approach is its conceptual compatibility with arbitrary forms of pathways. Most notably, enRoute works well with pathways that are manually created, as they are available in large, public pathway databases. We demonstrate enRoute with pathways from the well-established KEGG database and expression as well as copy number datasets from humans and mice with more than 1,000 experiments. We validate enRoute using case studies with domain experts, who used enRoute to explore data for glioblastoma multiforme in humans and a model of steatohepatitis in mice.
V. Pascucci, G. Scorzelli, B. Summa, P.-T. Bremer, A. Gyulassy, C. Christensen, S. Philip, S. Kumar.
“The ViSUS Visualization Framework,” In High Performance Visualization: Enabling Extreme-Scale Scientific Insight, Chapman and Hall/CRC Computational Science, Ch. 19, Edited by E. Wes Bethel and Hank Childs (LBNL) and Charles Hansen (UofU), Chapman and Hall/CRC, 2012.
Z. Peng, E. Grundy, R.S. Laramee, G. Chen, N. Croft.
“Mesh-Driven Vector Field Clustering and Visualization: An Image-Based Approach,” In IEEE Transactions on Visualization and Computer Graphics, 2011, Vol. 18, No. 2, pp. 283--298. February, 2012.
DOI: 10.1109/TVCG.2011.25
Vector field visualization techniques have evolved very rapidly over the last two decades, however, visualizing vector fields on complex boundary surfaces from computational flow dynamics (CFD) still remains a challenging task. In part, this is due to the large, unstructured, adaptive resolution characteristics of the meshes used in the modeling and simulation process. Out of the wide variety of existing flow field visualization techniques, vector field clustering algorithms offer the advantage of capturing a detailed picture of important areas of the domain while presenting a simplified view of areas of less importance. This paper presents a novel, robust, automatic vector field clustering algorithm that produces intuitive and insightful images of vector fields on large, unstructured, adaptive resolution boundary meshes from CFD. Our bottom-up, hierarchical approach is the first to combine the properties of the underlying vector field and mesh into a unified error-driven representation. The motivation behind the approach is the fact that CFD engineers may increase the resolution of model meshes according to importance. The algorithm has several advantages. Clusters are generated automatically, no surface parameterization is required, and large meshes are processed efficiently. The most suggestive and important information contained in the meshes and vector fields is preserved while less important areas are simplified in the visualization. Users can interactively control the level of detail by adjusting a range of clustering distance measure parameters. We describe two data structures to accelerate the clustering process. We also introduce novel visualizations of clusters inspired by statistical methods. We apply our method to a series of synthetic and complex, real-world CFD meshes to demonstrate the clustering algorithm results.
Keywords: Vector Field Visualization, Clustering, Feature-based, Surfaces
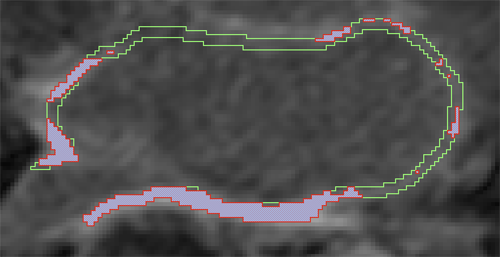
D. Perry, A. Morris, N. Burgon, C. McGann, R.S. MacLeod, J. Cates.
“Automatic classification of scar tissue in late gadolinium enhancement cardiac MRI for the assessment of left-atrial wall injury after radiofrequency ablation,” In SPIE Proceedings, Vol. 8315, pp. (published online). 2012.
DOI: 10.1117/12.910833
PubMed ID: 24236224
PubMed Central ID: PMC3824273
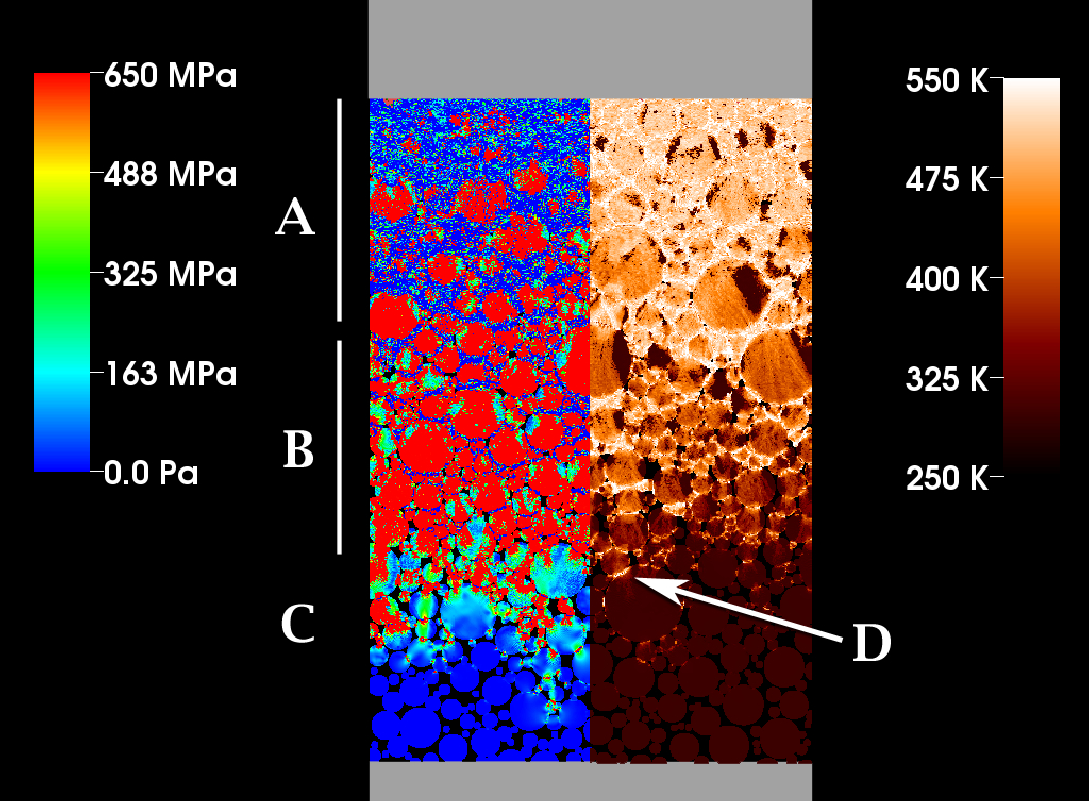
J.R. Peterson, J.C. Beckvermit, T. Harman, M. Berzins, C.A. Wight.
“Multiscale Modeling of High Explosives for Transportation Accidents,” In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond, 2012.
DOI: 10.1145/2335755.2335828
J. Peterson, C. Wight.
“An Eulerian-Lagrangian Computational Model for Deagration and Detonation of High Explosives,” In Combustion and Flame, Vol. 159, No. 7, pp. 2491--2499. 2012.
DOI: 10.1016/j.combustflame.2012.02.006
A solid phase explosives deflagration and detonation model capable of surface burning, convective bulk burning and detonation is formulated in the context of Eulerian–Lagrangian material mechanics. Well-validated combustion and detonation models, WSB and JWL++, are combined with two simple, experimentally indicated transition thresholds partitioning the three reaction regimes. Standard experiments are simulated, including the Aluminum Flyer Plate test, the Cylinder test, the rate stick test and the Steven test in order to validate the model. Cell and particle resolution dependence of simulation metrics are presented and global uncertainties assigned. Error quantification comparisons with experiments led to values generally below 7% (1σ). Finally, gas flow through porous media is implicated as the driving force behind the deflagration to detonation transition.
S.P. Ponnapalli, M.A. Saunders, C.F. Van Loan, O. Alter.
“A Higher-Order Generalized Singular Value Decomposition for Comparison of Global mRNA Expression from Multiple Organisms,” In PLoS One, Vol. 6, No. 12, pp. e28072. 2012.
DOI: 10.1371/journal.pone.0028072
K. Potter, R.M. Kirby, D. Xiu, C.R. Johnson.
“Interactive visualization of probability and cumulative density functions,” In International Journal of Uncertainty Quantification, Vol. 2, No. 4, pp. 397--412. 2012.
DOI: 10.1615/Int.J.UncertaintyQuantification.2012004074
PubMed ID: 23543120
PubMed Central ID: PMC3609671
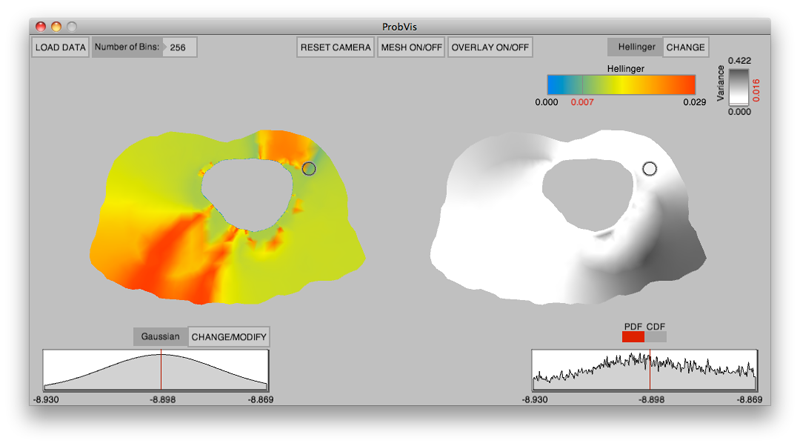
Keywords: visualization, probability density function, cumulative density function, generalized polynomial chaos, stochastic Galerkin methods, stochastic collocation methods
K. Potter, P. Rosen, C.R. Johnson.
“From Quantification to Visualization: A Taxonomy of Uncertainty Visualization Approaches,” In Uncertainty Quantification in Scientific Computing, IFIP Advances in Information and Communication Technology Series, Vol. 377, Edited by Andrew Dienstfrey and Ronald Boisvert, Springer, pp. 226--249. 2012.
DOI: 10.1007/978-3-642-32677-6_15
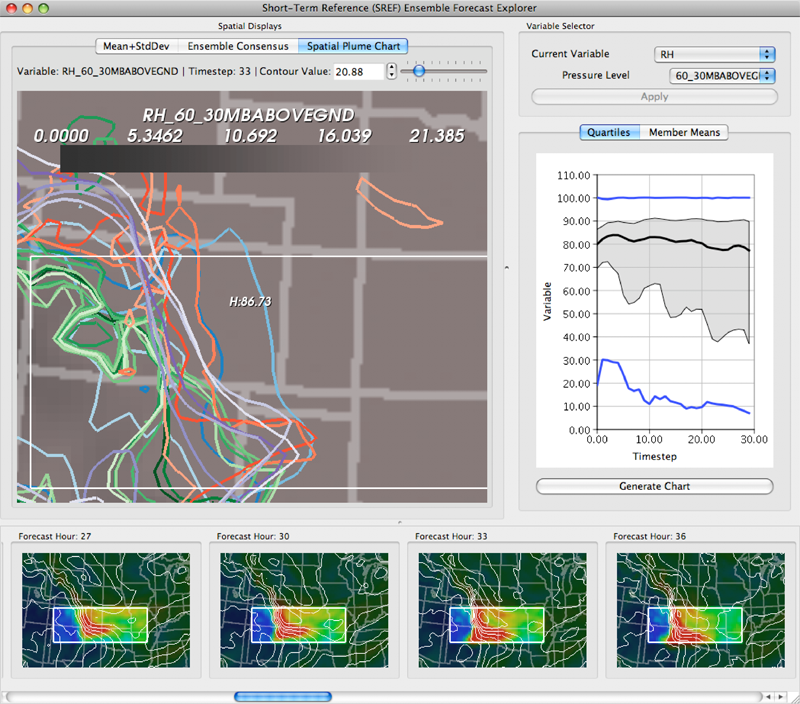
Keywords: scidac, netl, uncertainty visualization
M.W. Prastawa, S.P. Awate, G. Gerig.
“Building Spatiotemporal Anatomical Models using Joint 4-D Segmentation, Registration, and Subject-Speci fic Atlas Estimation,” In Proceedings of the 2012 IEEE Mathematical Methods in Biomedical Image Analysis (MMBIA) Conference, pp. 49--56. 2012.
DOI: 10.1109/MMBIA.2012.6164740
PubMed ID: 23568185
PubMed Central ID: PMC3615562
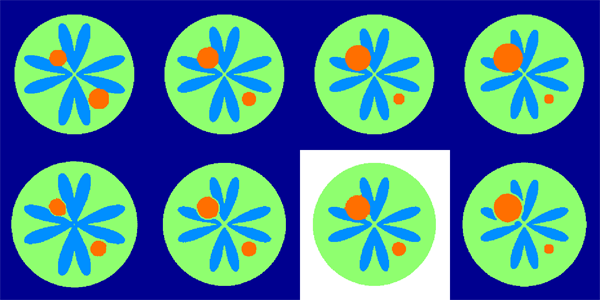
Keywords: namic, adni, autism
R. Pulch, D. Xiu.
“Generalised Polynomial Chaos for a Class of Linear Conservation Laws,” In Journal of Scientific Computing, Vol. 51, No. 2, pp. 293--312. 2012.
DOI: 10.1007/s10915-011-9511-5
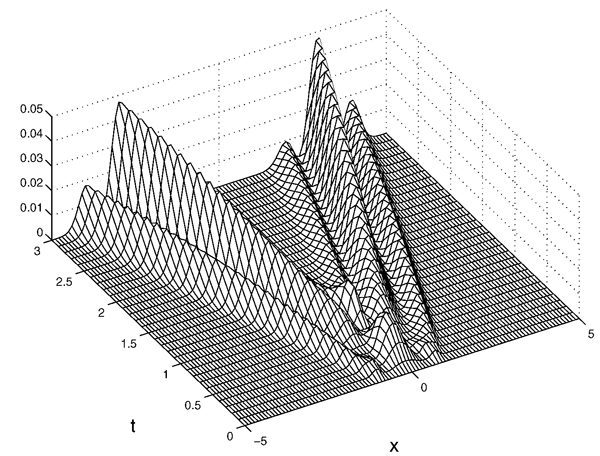
Keywords: Generalised polynomial chaos, Galerkin method, Random parameter, Conservation laws, Hyperbolic systems
N. Ramesh, B. J. Dangott, M. Salama, T. Tasdizen.
“Segmentation and Two-Step Classification of White Blood Cells in Peripheral Blood Smear,” In Journal of Pathology Informatics, Vol. 3, No. 13, 2012.
An automated system for differential white blood cell (WBC) counting based on morphology can make manual differential leukocyte counts faster and less tedious for pathologists and laboratory professionals. We present an automated system for isolation and classification of WBCs in manually prepared, Wright stained, peripheral blood smears from whole slide images (WSI). Methods: A simple, classification scheme using color information and morphology is proposed. The performance of the algorithm was evaluated by comparing our proposed method with a hematopathologist's visual classification. The isolation algorithm was applied to 1938 subimages of WBCs, 1804 of them were accurately isolated. Then, as the first step of a two-step classification process, WBCs were broadly classified into cells with segmented nuclei and cells with nonsegmented nuclei. The nucleus shape is one of the key factors in deciding how to classify WBCs. Ambiguities associated with connected nuclear lobes are resolved by detecting maximum curvature points and partitioning them using geometric rules. The second step is to define a set of features using the information from the cytoplasm and nuclear regions to classify WBCs using linear discriminant analysis. This two-step classification approach stratifies normal WBC types accurately from a whole slide image. Results: System evaluation is performed using a 10-fold cross-validation technique. Confusion matrix of the classifier is presented to evaluate the accuracy for each type of WBC detection. Experiments show that the two-step classification implemented achieves a 93.9\% overall accuracy in the five subtype classification. Conclusion: Our methodology achieves a semiautomated system for the detection and classification of normal WBCs from scanned WSI. Further studies will be focused on detecting and segmenting abnormal WBCs, comparison of 20x and 40x data, and expanding the applications for bone marrow aspirates.
Page 55 of 144
