 Despite great advances in neuroscience and medical technology in recent decades, nearly ten million Americans still suffer blindness due to retinal degenerative diseases such as retinitis pigmentosa (RP), age-related macular degeneration (AMD), diabetic retinopathy, and glaucoma. Unfortunately, current treatments available for these conditions are still quite limited. A primary challenge to developing effective treatments is the need for a complete understanding of the highly complex and delicate systems that compose the retina and how those systems change in response to degenerative disorders.
Despite great advances in neuroscience and medical technology in recent decades, nearly ten million Americans still suffer blindness due to retinal degenerative diseases such as retinitis pigmentosa (RP), age-related macular degeneration (AMD), diabetic retinopathy, and glaucoma. Unfortunately, current treatments available for these conditions are still quite limited. A primary challenge to developing effective treatments is the need for a complete understanding of the highly complex and delicate systems that compose the retina and how those systems change in response to degenerative disorders.Remodeling processes that occur in the neuronal pathways within the retina during the course of retinal deterioration are of particular importance to the development of treatments for these conditions. Researchers at the Robert E. Marc Laboratory at the Moran Eye Center are collaborating with the SCI Institute on a project supported by the NIH-NIBIB (grant number 5R01EB005832) to develop high-throughput techniques for reconstructing and visualizing the neural structures that compose the retina in order to meet these challenges.
Scientific Background
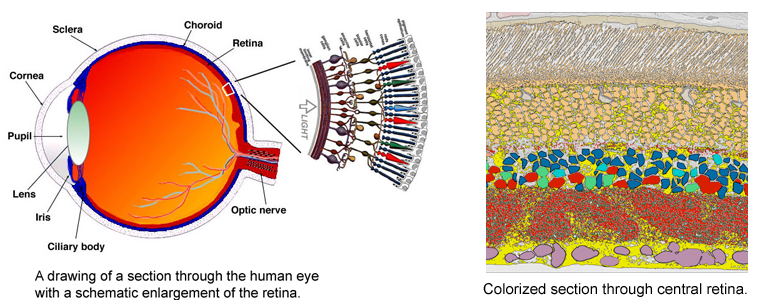
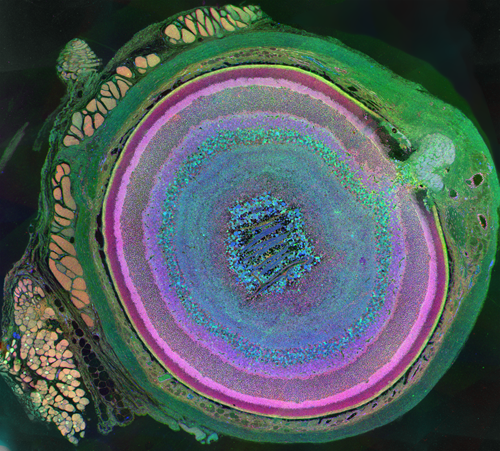
The very thin layer of cells at the back of the eye that comprise the retina do more than just translate light into nerve signals that are transmitted to the brain. The retina is actually a layer cake of highly specialized cells that perform a great deal of image processing and signal amplification before sending the signals through the optic nerve to the brain. Over the past eight years, the MarcLab has shown that blinding retinal diseases such as retinitis pigmentosa (RP) and age-related macular degeneration (AMD) trigger pathological rewiring patterns in the retina, as though the retina had acquired epilepsy or Alzheimer's and was now unable to process image data. This meant that schemes like transplants or bionic implants would not work in most patients. Neuroscientists will have to learn how to stop the rewiring, work around it, re-direct it or even replace the entire retina using synthetic biology. Using knowledge of how a real retina is built in its finest details (not just guessing) will guide how they might build a synthetic eye from biological sources. Neural circuit reconstruction and imaging are key tools in this effort.
"The aims of our laboratory include developing (I) a complete catalogue of retinal neurons, (II) a complete map of their connections, and (III) a detailed description of how networks respond to blinding diseases. The long-term goals are to find out how neuron networks shape what we see and how those networks break down in disease.
These goals require two tool sets: (I) rich libraries of molecular tags to identify cells (Marc Lab) and (II) imaging methods to track their forms, spatial patterns and connections (SCI). A human retina could contain over 10,000 unique network types but we have shown it uses only about 20 network types. But which ones? This can only be determined by tracing the connections of identified network partners. Then we can track changes in identified networks in genetic disease models." -- Dr. Robert Marc (Marc Lab Director)
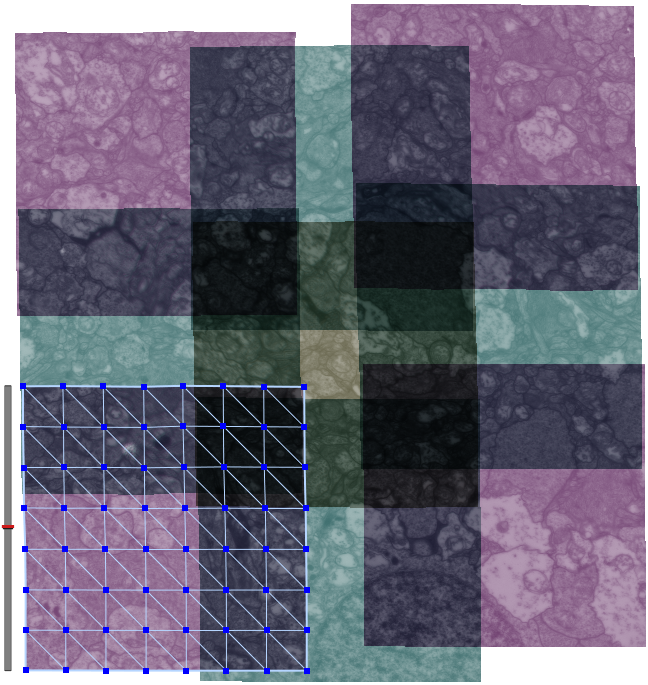
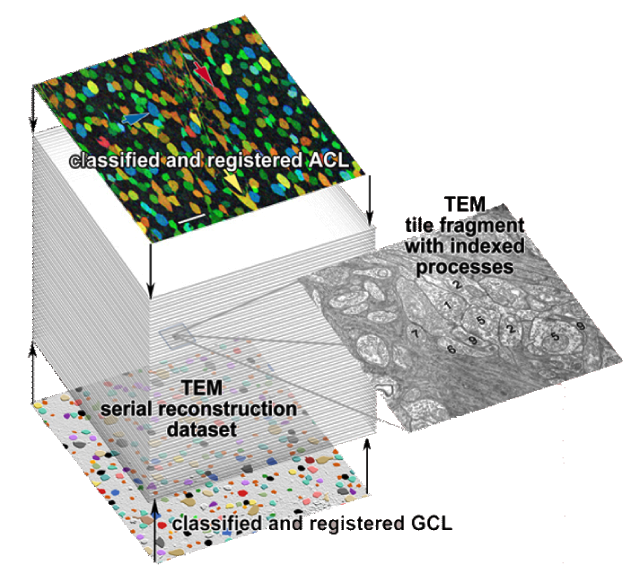
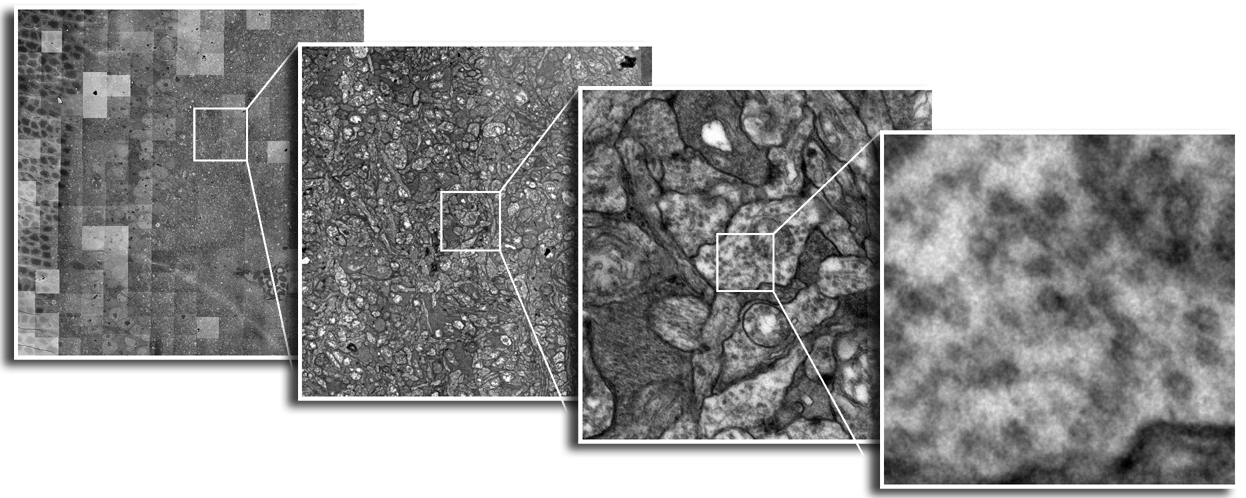
Until recently, methods used to reconstruct the structure of the retina were time consuming and labor intensive. The standard process known as serial-section transmission electron microscopy (TEM) involves slicing a sample of retinal tissue into a series of extremely thin layers and then imaging each slice with an electron microscope. The slices are cut with a diamond knive with incredible precision, as thin as 1/1000 the thickness of a sheet of newspaper. Due to the limited field of view delivered by the microscope it is necessary to take many images, each covering a small patch of the slice, and then reassemble the 'tiles' in software to build a complete mosaic image of each slice. The scientist would then have to manually trace around the neurons in each slice and attempt to follow them through successive layers in order to reconstruct the connectivities of the neurons. The manual nature of this process meant that while it was possible to construct a representation of a small section of an example retina, it was not practical to do so for many examples in a variety of conditions or track changes that occur over the course of a disease.
Software Tools
SCI Institute scientists have developed a suite of software tools to automate the process of building TEM image mosaics and assembling the slices into 3D volumes. The speed and accuracy made possible through these systems has not only made it possible to reconstruct more retinal samples more quickly, but has also greatly expanded the level of detail possible. Marc Lab scientists are now assembling mosaics of several hundred individual TEM images in a single slice. Future targets of over 1000 tile mosaics are anticipated to be reached soon. While current volumes are composed of dozens of slices, future samples will be composed of hundreds of slices and require terabytes of storage space."The command-line mosaicing tools amaze me. These tools take a series of tiles as input and then assemble the tiles together like a puzzle to create a larger mosaic. This is a task that we used to do by hand. The SCI code automatically produces results which are nearly flawless. The tools were originally designed and tested to work on film electron microscopy (EM) images. They work as advertised to build large 200-tile (4096x4096 pixels per tile) EM datasets but they've been robust enough to mosaic light images as well with no modification. Before SCI we built light microscopy mosaics using a Syncroscan system that cost many tens of thousands of dollars. The Syncroscan mosaics are good, but mechanical issues result in each tile being placed several pixels away from its true position. I've now switched to using the SCI tools to build light microscopy mosaics since they produce nearly flawless results. Having the tools work that well on a task outside of the type of image they were tested on is a real testament to their quality. This need to build mosaics from individual tiles is a capability that many biological labs would love to have a cheap accessible solution for and I think the SCI tools meet that need." -- James Anderson (Marc Lab Graduate Student)
"The collaboration with the Marc Laboratory provides and excellent application to drive the development of this software, however we're also keeping an eye on the possibilities that exist for the broader scope of neuroscience. An important aspect of neurobiology is to decipher the patterns of neuronal connections that govern vertebrate behavior. Understanding brain function at the cellular level requires a detailed analysis of connectivity of individual neurons in regions that consist of densely packed cells and processes. Our team at the Scientific Computing and Imaging Institute is working towards innovative algorithms and tools to automate neural circuit reconstruction research as much as possible. We can see that the same technologies that have proven to be useful for retina research could easily be applied to a great number of other problems in neuroscience." -- Dr. Tolga Tasdizen (SCI Institute Principal Investigator)
The large datasets involved pose an additional challenge for SCI Software developers. Loading and manipulating such large datasets given the practical limitations of a typical laboratory PC requires new techniques for data interaction. SCI Programmers have employed a novel approach in a tool called Mosaic Builder whereby microscopy tiles are stored individually and a separate file records the transformations required to assemble them into the mosaics. The system can therefore dynamically reconstruct and display just the parts of the dataset needed at a given time. This method allows the software to handle arbitrarily large datasets.
"Mosaic Builder is the first SCI viewer we've started using to examine our electron microscopy mosaics. The command-line tools produce a text file containing the mathematical transforms required to mosaic the tiles. Mosaic Builder uses those transforms and efficient usage of memory to quickly display extremely large 4GB+ mosaics. Images of this size are very unwieldy in applications such as Photoshop. Mosaic Builder also has a built-in markup capability that allows us to go through a dataset and annotate structures of interest such as synapses. Previously markup was done using a text layer in Photoshop, so having points of interest along with a specific coordinate and meta-data is definitely a step up for us." -- James Anderson (Marc Lab)
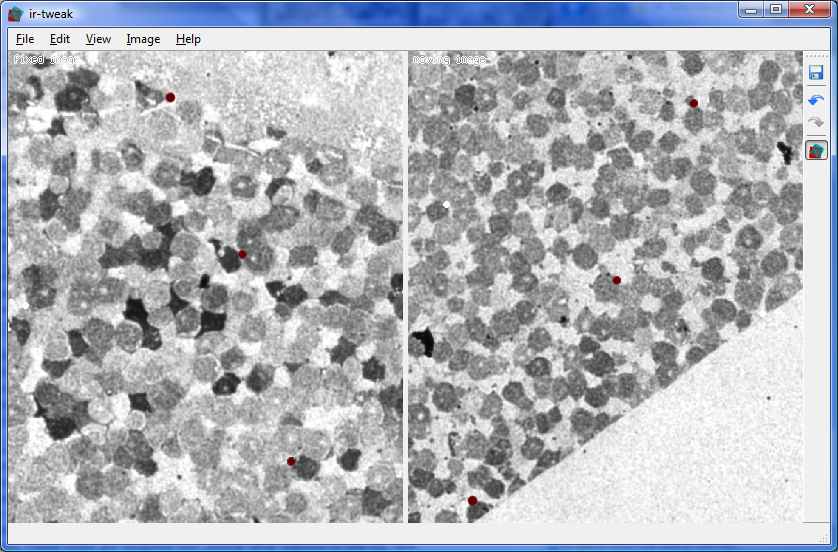
Another tool called ir-tweak assists in the process of matching up overlapping portions of adjacent image tiles to reconstruct the larger mosaics if the command-line tools fail. The matching process is complicated by the fact that small distortion effects in the microscope can warp portions of the images slighly and heat produced by the microscope can distort the samples themselves. Ir-tweak allows the user to re-warp portions of the images to compensate for this phenomenon and achieve much better results.
"We use ir-tweak to do registration when we need to align serial sections of tissue with different stains. Automatic solutions have not worked on this problem. SCI wrote us a GUI which allows us to interactively place control points on both images and view the resulting transform to see how well the two images are aligned. Once aligned the warped image can be exported. This was a big improvement from the old PCI Geomatica code which did not produce any dynamic feedback. Ir-tweak increased our throughput for registration dramatically." -- James Anderson (Marc Lab)
 |
 |
| Ir-tweak | Neuron Path Viewer |
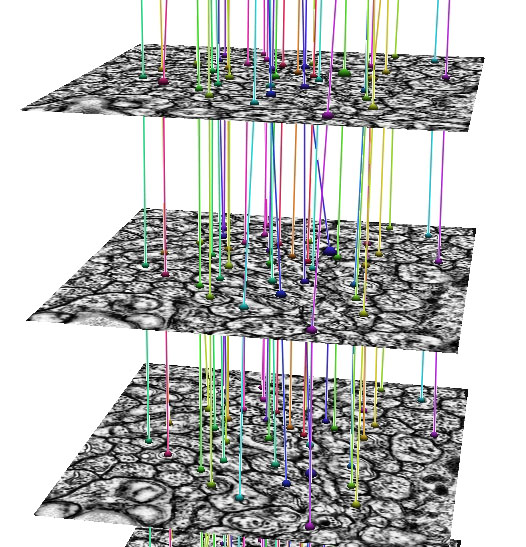
In development, is an automated Neuron Path Viewer to extract neuron connectivity and structure over large volumes of retina tissue. The current method formulates the problem as an optimal path finding algorithm through sets of sections allowing for variability and inconsistencies within the data. Neurons in each slice are segmented and costs are computed for segmented regions between slices that best represent the probability of a neuron match across a section. These costs are weighted with several other features to find the best sequence of regions that make up a neuron.
"The grazed membranes and extreme anisotropic nature of the data require us to use a strategy that accounts for large changes in the neuron shape, size and appearance. The optimal path finding method can take large changes in the data into account and choose the best representation of a neuron's structure through the volume." -- Elizabeth Jurrus (School of Computing PhD graduate student)
"SCI Institute tools improved three areas substantially. First, we needed statistically robust data samples, which meant building large image mosaics from tiles. SCI's MosaicBuilder is a major throughput advance, converting year-long data acquisition projects into a few days of automated tiling. Second, building a cell's molecular signature required precise multimodal image registration. SCI's ir-tweak is the best user-guided tool we have found in over 20 hears of image registration work. Our biggest needs will be tracking thousands of identified processes through complex networks to find the 20 right patterns that human vision requires. The SCI collaboration offers the hope that decoding networks and connections can be done in weeks to months rather than many years ... or never." -- Dr. Robert Marc (Marc Lab Director)
The collaboration of SCI computer scientists with Marc Laboratory neuroscientists has achieved an important advancement in the way retinal neuroscience research is conducted. The ability to quickly produce detailed computer models of retinal tissue samples and analyze the complex neural networks that make the human visual system function has opened new doorways in the fight against degenerative eye disease. Such advances bring us significantly closer to the time when blindness will be a thing of the past.
 |
 |
| SCI Institute: (Top L-R) Kannan UV, Dr. Tolga Tasdizen, Dr. Ross Whitaker. (bottom) Liz Jurrus, Joël Spaltenstein | Marc Lab: (L-R) Dr. Robert Marc, Dr. Bryan Jones, James R. Anderson |
Principal Researchers
SCI Institute- Dr. Tolga Tasdizen
- Dr. Ross T. Whitaker
- Joël Spaltenstein
- Elizabeth Jurrus
- Kannan U. V.
- Dr. Robert Marc
- Dr. Bryan Jones
- James R. Anderson
- Dr. Chi-Bin Chien
- Melissa Hardy