SCI Publications
2014
B. Chapman, H. Calandra, S. Crivelli, J. Dongarra, J. Hittinger, C.R. Johnson, S.A. Lathrop, V. Sarkar, E. Stahlberg, J.S. Vetter, D. Williams.
“ASCAC Workforce Subcommittee Letter,” Note: Office of Scientific and Technical Information, DOE ASCAC Committee Report, July, 2014.
DOI: 10.2172/1222711
Simulation and computing are essential to much of the research conducted at the DOE national laboratories. Experts in the ASCR-relevant Computing Sciences, which encompass a range of disciplines including Computer Science, Applied Mathematics, Statistics and domain sciences, are an essential element of the workforce in nearly all of the DOE national laboratories. This report seeks to identify the gaps and challenges facing DOE with respect to this workforce.
The DOE laboratories provided the committee with information on disciplines in which they experienced workforce gaps. For the larger laboratories, the majority of the cited workforce gaps were in the Computing Sciences. Since this category spans multiple disciplines, it was difficult to obtain comprehensive information on workforce gaps in the available timeframe. Nevertheless, five multi-purpose laboratories provided additional relevant data on recent hiring and retention.
Data on academic coursework was reviewed. Studies on multidisciplinary education in Computational Science and Engineering (CS&E) revealed that, while the number of CS&E courses offered is growing, the overall availability is low and the coursework fails to provide skills for applying CS&E to real-world applications. The number of graduates in different fields within Computer Science (CS) and Computer Engineering (CE) was also reviewed, which confirmed that specialization in DOE areas of interest is less common than in many other areas.
Projections of industry needs and employment figures (mostly for CS and CE) were examined. They indicate a high and increasing demand for graduates in all areas of computing, with little unemployment. This situation will be exacerbated by large numbers of retirees in the coming decade. Further, relatively few US students study toward higher degrees in the Computing Sciences, and those who do are predominantly white and male. As a result of this demographic imbalance, foreign nationals are an increasing fraction of the graduate population and we fail to benefit from including women and underrepresented minorities.
There is already a program that supports graduate education that is tailored to the needs of the DOE laboratories. The Computational Science Graduate Fellowship (CSGF) enables graduates to pursue a multidisciplinary program of education that is coupled with practical experience at the laboratories. It has been demonstrated to be highly effective in both its educational goals and in its ability to supply talent to the laboratories. However, its current size and scope are too limited to solve the workforce problems identified. The committee felt strongly that this proven program should be extended to increase its ability to support the DOE mission.
Since no single program can eliminate the workforce gap, existing recruitment efforts by the laboratories were examined. It was found that the laboratories already make considerable effort to recruit in this area. Although some challenges, such as the inability to match industry compensation, cannot be directly addressed, DOE could develop a roadmap to increase the impact of individual laboratory efforts, to enhance the suitability of existing educational opportunities, to increase the attractiveness of the laboratories, and to attract and sustain a full spectrum of human talent, which includes women and underrepresented minorities.
S.E. Cooper, K.G. Driesslein, A.M. Noecker, C.C. McIntyre, A.M. Machado, C.R. Butson.
“Anatomical targets associated with abrupt versus gradual washout of subthalamic deep brain stimulation effects on bradykinesia,” In PloS One, Vol. 9, No. 8, pp. e99663. January, 2014.
ISSN: 1932-6203
DOI: 10.1371/journal.pone.0099663
PubMed ID: 25098453

A. Dubey, A. Almgren, John Bell, M. Berzins, S. Brandt, G. Bryan, P. Colella, D. Graves, M. Lijewski, F. Löffler, B. O’Shea, E. Schnetter, B. Van Straalen, K. Weide.
“A survey of high level frameworks in block-structured adaptive mesh refinement packages,” In Journal of Parallel and Distributed Computing, 2014.
DOI: 10.1016/j.jpdc.2014.07.001
Over the last decade block-structured adaptive mesh refinement (SAMR) has found increasing use in large, publicly available codes and frameworks. SAMR frameworks have evolved along different paths. Some have stayed focused on specific domain areas, others have pursued a more general functionality, providing the building blocks for a larger variety of applications. In this survey paper we examine a representative set of SAMR packages and SAMR-based codes that have been in existence for half a decade or more, have a reasonably sized and active user base outside of their home institutions, and are publicly available. The set consists of a mix of SAMR packages and application codes that cover a broad range of scientific domains. We look at their high-level frameworks, their design trade-offs and their approach to dealing with the advent of radical changes in hardware architecture. The codes included in this survey are BoxLib, Cactus, Chombo, Enzo, FLASH, and Uintah.
Keywords: SAMR, BoxLib, Chombo, FLASH, Cactus, Enzo, Uintah
S. Durrleman, M. Prastawa, N. Charon, J.R. Korenberg, S. Joshi, G. Gerig, A. Trouvé.
“Morphometry of anatomical shape complexes with dense deformations and sparse parameters,” In NeuroImage, 2014.
DOI: 10.1016/j.neuroimage.2014.06.043
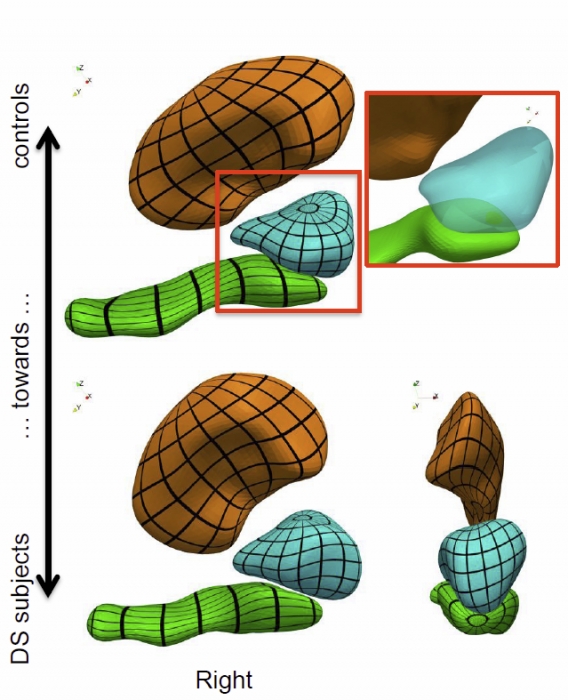
We propose a generic method for the statistical analysis of collections of anatomical shape complexes, namely sets of surfaces that were previously segmented and labeled in a group of subjects. The method estimates an anatomical model, the template complex, that is representative of the population under study. Its shape reflects anatomical invariants within the dataset. In addition, the method automatically places control points near the most variable parts of the template complex. Vectors attached to these points are parameters of deformations of the ambient 3D space. These deformations warp the template to each subject’s complex in a way that preserves the organization of the anatomical structures. Multivariate statistical analysis is applied to these deformation parameters to test for group differences. Results of the statistical analysis are then expressed in terms of deformation patterns of the template complex, and can be visualized and interpreted.
The user needs only to specify the topology of the template complex and the number of control points. The method then automatically estimates the shape of the template complex, the optimal position of control points and deformation parameters. The proposed approach is completely generic with respect to any type of application and well adapted to efficient use in clinical studies, in that it does not require point correspondence across surfaces and is robust to mesh imperfections such as holes, spikes, inconsistent orientation or irregular meshing.
The approach is illustrated with a neuroimaging study of Down syndrome (DS). Results demonstrate that the complex of deep brain structures shows a statistically significant shape difference between control and DS subjects. The deformation-based modelingis able to classify subjects with very high specificity and sensitivity, thus showing important generalization capability even given a low sample size. We show that results remain significant even if the number of control points, and hence the dimension of variables in the statistical model, are drastically reduced. The analysis may even suggest that parsimonious models have an increased statistical performance.
The method has been implemented in the software Deformetrica, which is publicly available at www.deformetrica.org.
Keywords: morphometry, deformation, varifold, anatomy, shape, statistics
L.T. Edgar, C.J. Underwood, J.E. Guilkey, J.B. Hoying, J.A. Weiss.
“Extracellular matrix density regulates the rate of neovessel growth and branching in sprouting angiogenesis,” In PLOS one, Vol. 9, No. 1, 2014.
DOI: 10.1371/journal.pone.0085178
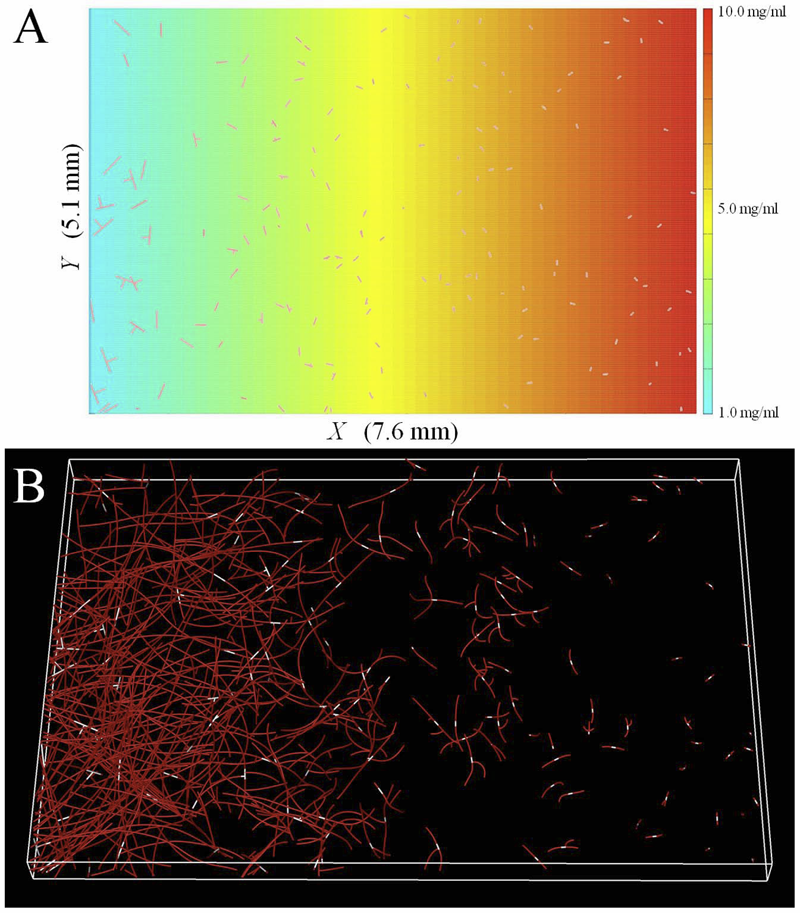
S. Elhabian, Y. Gur, C. Vachet, J. Piven, M. Styner, I. Leppert, G.B. Pike, G. Gerig.
“A Preliminary Study on the Effect of Motion Correction On HARDI Reconstruction,” In Proceedings of the 2014 IEEE International Symposium on Biomedical Imaging (ISBI), pp. (accepted). 2014.
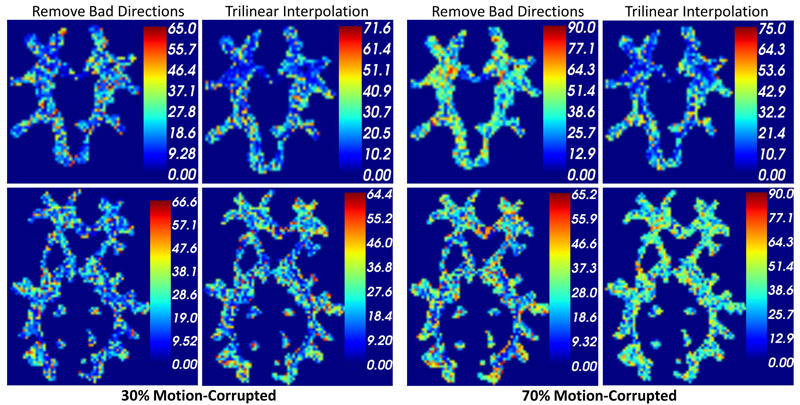
Keywords: Diffusion MRI, HARDI, motion correction, interpolation
S. Elhabian, Y. Gur, J. Piven, M. Styner, I. Leppert, G.B. Pike, G. Gerig.
“Subject-Motion Correction in HARDI Acquisitions: Choices and Consequences,” In Proceeding of the 2014 Joint Annual Meeting ISMRM-ESMRMB, pp. (accepted). 2014.
DOI: 10.3389/fneur.2014.00240
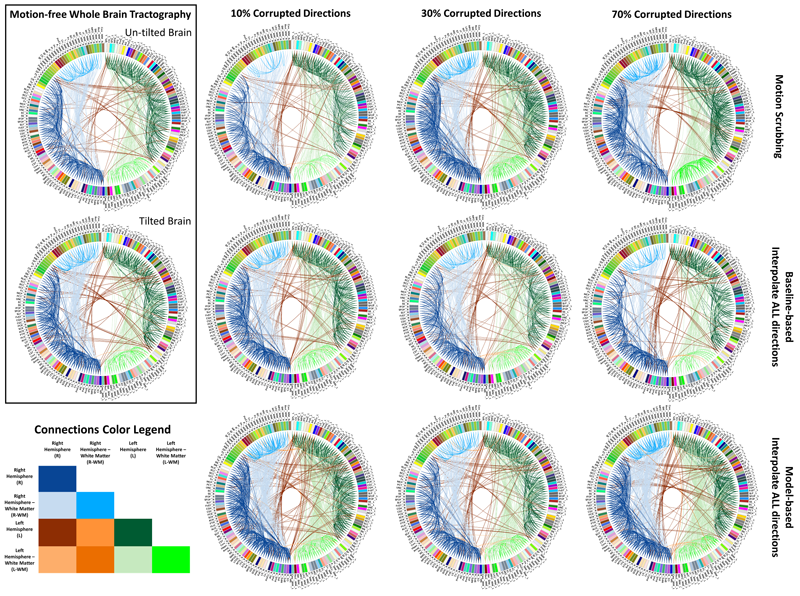
Unlike anatomical MRI where subject motion can most often be assessed by quick visual quality control, the detection, characterization and evaluation of the impact of motion in diffusion imaging are challenging issues due to the sensitivity of diffusion weighted imaging (DWI) to motion originating from vibration, cardiac pulsation, breathing and head movement. Post-acquisition motion correction is widely performed, e.g. using the open-source DTIprep software [1,2] or TORTOISE [3], but in particular in high angular resolution diffusion imaging (HARDI), users often do not fully understand the consequences of different types of correction schemes on the final analysis, and whether those choices may introduce confounding factors when comparing populations. Although there is excellent theoretical work on the number of directional DWI and its effect on the quality and crossing fiber resolution of orientation distribution functions (ODF), standard users lack clear guidelines and recommendations in practical settings. This research investigates motion correction using transformation and interpolation of affected DWI directions versus the exclusion of subsets of DWI’s, and its effects on diffusion measurements on the reconstructed fiber orientation diffusion functions and on the estimated fiber orientations. The various effects are systematically studied via a newly developed synthetic phantom and also on real HARDI data.
S. Elhabian, Y. Gur, J. Piven, M. Styner, I. Leppert, G. Bruce Pike, G. Gerig.
“Motion is inevitable: The impact of motion correction schemes on hardi reconstructions,” In Proceedings of the MICCAI 2014 Workshop on Computational Diffusion MRI, September, 2014.
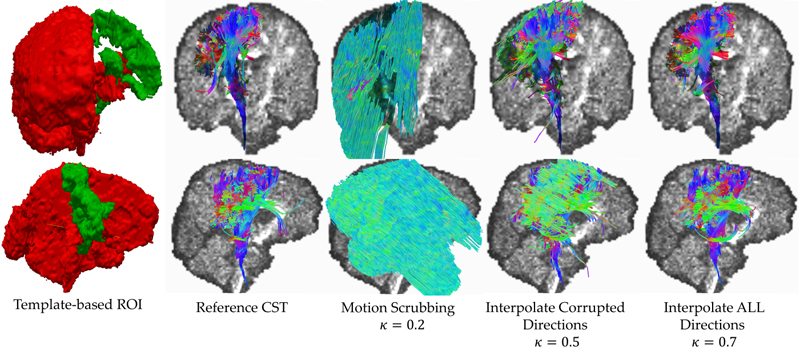
S.Y. Elhabian, Y. Gur, C. Vachet, J. Piven, M.A. Styner, I.R. Leppert, B. Pike, G. Gerig.
“Subject-Motion Correction in HARDI Acquisitions: Choices and Consequences,” In Frontiers in Neurology - Brain Imaging Methods, 2014.
DOI: 10.3389/fneur.2014.00240
T. Etiene, D. Jonsson, T. Ropinski, C. Scheidegger, J.L.D. Comba, L. G. Nonato, R. M. Kirby, A. Ynnerman,, C. T. Silva.
“Verifying Volume Rendering Using Discretization Error Analysis,” In IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS, Vol. 20, No. 1, IEEE, pp. 140-154. January, 2014.
We propose an approach for verification of volume rendering correctness based on an analysis of the volume rendering integral, the basis of most DVR algorithms. With respect to the most common discretization of this continuous model (Riemann summation), we make assumptions about the impact of parameter changes on the rendered results and derive convergence curves describing the expected behavior. Specifically, we progressively refine the number of samples along the ray, the grid size, and the pixel size, and evaluate how the errors observed during refinement compare against the expected approximation errors. We derive the theoretical foundations of our verification approach, explain how to realize it in practice, and discuss its limitations. We also report the errors identified by our approach when applied to two publicly available volume rendering packages.
A. Faucett, T. Harman, T. Ameel.
“Computational Determination of the Modified Vortex Shedding Frequency for a Rigid, Truncated, Wall-Mounted Cylinder in Cross Flow,” In Volume 10: Micro- and Nano-Systems Engineering and Packaging, Montreal, ASME International Mechanical Engineering Congress and Exposition (IMECE), International Conference on Computational Science, November, 2014.
DOI: 10.1115/imece2014-39064
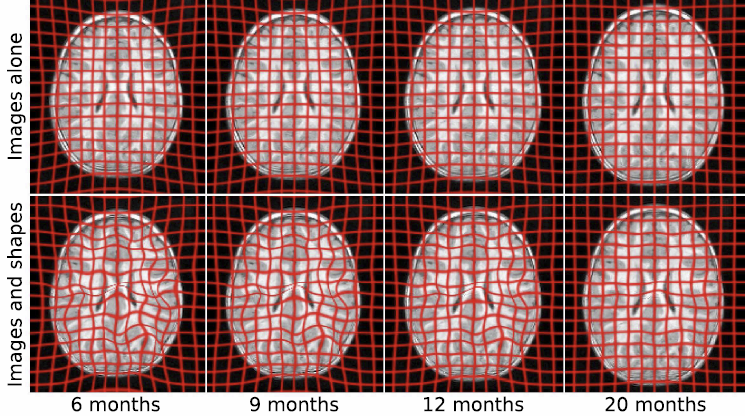
J. Fishbaugh, M. Prastawa, G. Gerig, S. Durrleman.
“Geodesic Regression of Image and Shape Data for Improved Modeling of 4D Trajectories,” In Proceedings of the 2014 IEEE International Symposium on Biomedical Imaging (ISBI), pp. (accepted). 2014.
T. Fogal, F. Proch, A. Schiewe, O. Hasemann, A. Kempf, J. Krüger.
“Freeprocessing: Transparent in situ visualization via data interception,” In Proceedings of the 14th Eurographics Conference on Parallel Graphics and Visualization, EGPGV, Eurographics Association, 2014.
In situ visualization has become a popular method for avoiding the slowest component of many visualization pipelines: reading data from disk. Most previous in situ work has focused on achieving visualization scalability on par with simulation codes, or on the data movement concerns that become prevalent at extreme scales. In this work, we consider in situ analysis with respect to ease of use and programmability. We describe an abstraction that opens up new applications for in situ visualization, and demonstrate that this abstraction and an expanded set of use cases can be realized without a performance cost.
Z. Fu, H.K. Dasari, M. Berzins, B. Thompson.
“Parallel Breadth First Search on GPU Clusters,” SCI Technical Report, No. UUSCI-2014-002, SCI Institute, University of Utah, 2014.
Fast, scalable, low-cost, and low-power execution of parallel graph algorithms is important for a wide variety of commercial and public sector applications. Breadth First Search (BFS) imposes an extreme burden on memory bandwidth and network communications and has been proposed as a benchmark that may be used to evaluate current and future parallel computers. Hardware trends and manufacturing limits strongly imply that many core devices, such as NVIDIA® GPUs and the Intel® Xeon Phi®, will become central components of such future systems. GPUs are well known to deliver the highest FLOPS/watt and enjoy a very significant memory bandwidth advantage over CPU architectures. Recent work has demonstrated that GPUs can deliver high performance for parallel graph algorithms and, further, that it is possible to encapsulate that capability in a manner that hides the low level details of the GPU architecture and the CUDA language but preserves the high throughput of the GPU. We extend previous research on GPUs and on scalable graph processing on super-computers and demonstrate that a high-performance parallel graph machine can be created using commodity GPUs and networking hardware.
Keywords: GPU cluster, MPI, BFS, graph, parallel graph algorithm
Z. Fu, H.K. Dasari, M. Berzins, B. Thompson.
“Parallel Breadth First Search on GPU Clusters,” In Proceedings of the IEEE BigData 2014 Conference, Washington DC, October, 2014.
Fast, scalable, low-cost, and low-power execution of parallel graph algorithms is important for a wide variety of commercial and public sector applications. Breadth First Search (BFS) imposes an extreme burden on memory bandwidth and network communications and has been proposed as a benchmark that may be used to evaluate current and future parallel computers. Hardware trends and manufacturing limits strongly imply that many core devices, such as NVIDIA® GPUs and the Intel® Xeon Phi®, will become central components of such future systems. GPUs are well known to deliver the highest FLOPS/watt and enjoy a very significant memory bandwidth advantage over CPU architectures. Recent work has demonstrated that GPUs can deliver high performance for parallel graph algorithms and, further, that it is possible to encapsulate that capability in a manner that hides the low level details of the GPU architecture and the CUDA language but preserves the high throughput of the GPU. We extend previous research on GPUs and on scalable graph processing on super-computers and demonstrate that a high-performance parallel graph machine can be created using commodity GPUs and networking hardware.
Keywords: GPU cluster, MPI, BFS, graph, parallel graph algorithm
Y. Gao, M. Prastawa, M. Styner, J. Piven, G. Gerig.
“A Joint Framework for 4D Segmentation and Estimation of Smooth Temporal Appearance Changes,” In Proceedings of the 2014 IEEE International Symposium on Biomedical Imaging (ISBI), pp. (accepted). 2014.
M.G. Genton, C.R. Johnson, K. Potter, G. Stenchikov, Y. Sun.
“Surface boxplots,” In Stat Journal, Vol. 3, No. 1, pp. 1--11. 2014.
In this paper, we introduce a surface boxplot as a tool for visualization and exploratory analysis of samples of images. First, we use the notion of volume depth to order the images viewed as surfaces. In particular, we define the median image. We use an exact and fast algorithm for the ranking of the images. This allows us to detect potential outlying images that often contain interesting features not present in most of the images. Second, we build a graphical tool to visualize the surface boxplot and its various characteristics. A graph and histogram of the volume depth values allow us to identify images of interest. The code is available in the supporting information of this paper. We apply our surface boxplot to a sample of brain images and to a sample of climate model outputs.
T. Geymayer, M. Steinberger, A. Lex, M. Streit,, D. Schmalstieg.
“Show me the Invisible: Visualizing Hidden Content,” In Proceedings of the ACM SIGCHI Conference on Human Factors in Computing Systems (CHI '14), CHI '14, ACM, pp. 3705--3714. 2014.
ISBN: 978-1-4503-2473-1
DOI: 10.1145/2556288.2557032
Content on computer screens is often inaccessible to users because it is hidden, e.g., occluded by other windows, outside the viewport, or overlooked. In search tasks, the efficient retrieval of sought content is important. Current software, however, only provides limited support to visualize hidden occurrences and rarely supports search synchronization crossing application boundaries. To remedy this situation, we introduce two novel visualization methods to guide users to hidden content. Our first method generates awareness for occluded or out-of-viewport content using see-through visualization. For content that is either outside the screen's viewport or for data sources not opened at all, our second method shows off-screen indicators and an on-demand smart preview. To reduce the chances of overlooking content, we use visual links, i.e., visible edges, to connect the visible content or the visible representations of the hidden content. We show the validity of our methods in a user study, which demonstrates that our technique enables a faster localization of hidden content compared to traditional search functionality and thereby assists users in information retrieval tasks.
S. Gratzl, N. Gehlenborg, A. Lex, H. Pfister, M. Streit.
“Domino: Extracting, Comparing, and Manipulating Subsets across Multiple Tabular Datasets,” In IEEE Transactions on Visualization and Computer Graphics (InfoVis '14), Vol. 20, No. 12, pp. 2023--2032. 2014.
ISSN: 1077-2626
DOI: 10.1109/TVCG.2014.2346260
Answering questions about complex issues often requires analysts to take into account information contained in multiple interconnected datasets. A common strategy in analyzing and visualizing large and heterogeneous data is dividing it into meaningful subsets. Interesting subsets can then be selected and the associated data and the relationships between the subsets visualized. However, neither the extraction and manipulation nor the comparison of subsets is well supported by state-of-the-art techniques. In this paper we present Domino, a novel multiform visualization technique for effectively representing subsets and the relationships between them. By providing comprehensive tools to arrange, combine, and extract subsets, Domino allows users to create both common visualization techniques and advanced visualizations tailored to specific use cases. In addition to the novel technique, we present an implementation that enables analysts to manage the wide range of options that our approach offers. Innovative interactive features such as placeholders and live previews support rapid creation of complex analysis setups. We introduce the technique and the implementation using a simple example and demonstrate scalability and effectiveness in a use case from the field of cancer genomics.
K. Grewen, M. Burchinal, C. Vachet, S. Gouttard, J.H. Gilmore, W. Lin, J. Johns, M. Elam, G. Gerig.
“Prenatal cocaine effects on brain structure in early infancy,” In NeuroImage, Vol. 101, pp. 114--123. November, 2014.
DOI: 10.1016/j.neuroimage.2014.06.070
Page 39 of 144
