SCI Publications
2014
C.E. Gritton.
“Ringing Instabilities in Particle Methods,” Note: M.S. in Computational Engineering and Science, advisor Martin Berzins, School of Computing, University of Utah, August, 2014.
Particle methods have been used in fields ranging from fluid dynamics to plasma physics. The Particle-In-Cell method and the family of methods that are an extension of it are a combination of both Lagrangian and Eularian methods. In this thesis, we present a brief survey of some of the methods and their key components. We show the different methods by which spatial derviates are computed. We propose a method of showing how the so-called "ringing instabilies" associated with particle methods arise and a means to remove them. We also propose that the underlying nodal scheme plays a key role in the stability of the method. Lastly, different particle methods are explored through numerical simulations and compared against an analytic solution.
Y. Gur, C.R. Johnson.
“Generalized HARDI Invariants by Method of Tensor Contraction,” In Proceedings of the 2014 IEEE International Symposium on Biomedical Imaging (ISBI), pp. 718--721. April, 2014.
We propose a 3D object recognition technique to construct rotation invariant feature vectors for high angular resolution diffusion imaging (HARDI). This method uses the spherical harmonics (SH) expansion and is based on generating rank-1 contravariant tensors using the SH coefficients, and contracting them with covariant tensors to obtain invariants. The proposed technique enables the systematic construction of invariants for SH expansions of any order using simple mathematical operations. In addition, it allows construction of a large set of invariants, even for low order expansions, thus providing rich feature vectors for image analysis tasks such as classification and segmentation. In this paper, we use this technique to construct feature vectors for eighth-order fiber orientation distributions (FODs) reconstructed using constrained spherical deconvolution (CSD). Using simulated and in vivo brain data, we show that these invariants are robust to noise, enable voxel-wise classification, and capture meaningful information on the underlying white matter structure.
Keywords: Diffusion MRI, HARDI, invariants
C. Hamani, B.O. Amorim, A.L. Wheeler, M. Diwan, K. Driesslein, L. Covolan, C.R. Butson, J.N. Nobrega.
“Deep brain stimulation in rats: Different targets induce similar antidepressant-like effects but influence different circuits,” In Neurobiology of Disease, Vol. 71, Elsevier Inc., pp. 205--214. August, 2014.
ISSN: 1095-953X
DOI: 10.1016/j.nbd.2014.08.007
PubMed ID: 25131446
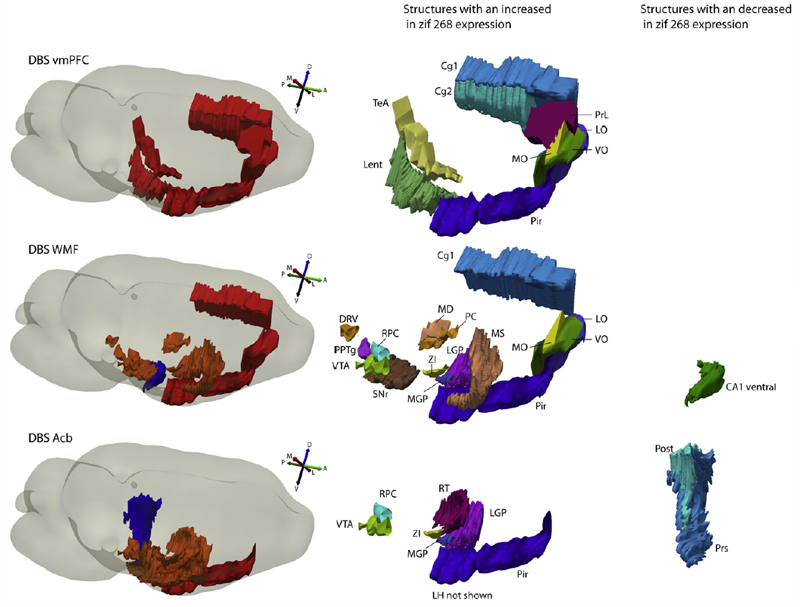
Keywords: Anterior capsule, Connectivity, Deep brain stimulation, Depression, Nucleus accumbens, Prefrontal cortex
C.D. Hansen, M. Chen, C.R. Johnson, A.E. Kaufman, H. Hagen (Eds.).
“Scientific Visualization: Uncertainty, Multifield, Biomedical, and Scalable Visualization,” Mathematics and Visualization, Springer, 2014.
ISBN: 978-1-4471-6496-8
X. Hao, K. Zygmunt, R.T. Whitaker, P.T. Fletcher.
“Improved Segmentation of White Matter Tracts with Adaptive Riemannian Metrics,” In Medical Image Analysis, Vol. 18, No. 1, pp. 161--175. Jan, 2014.
DOI: 10.1016/j.media.2013.10.007
PubMed ID: 24211814
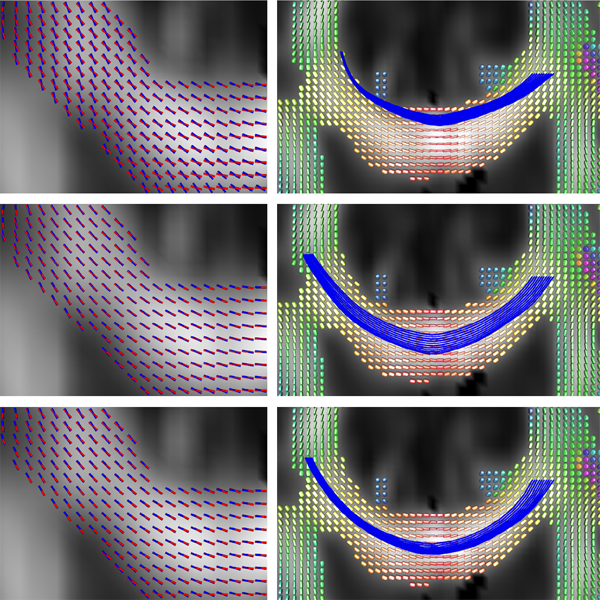
Keywords: Conformal factor, Diffusion tensor imaging, Front-propagation, Geodesic, Riemannian manifold
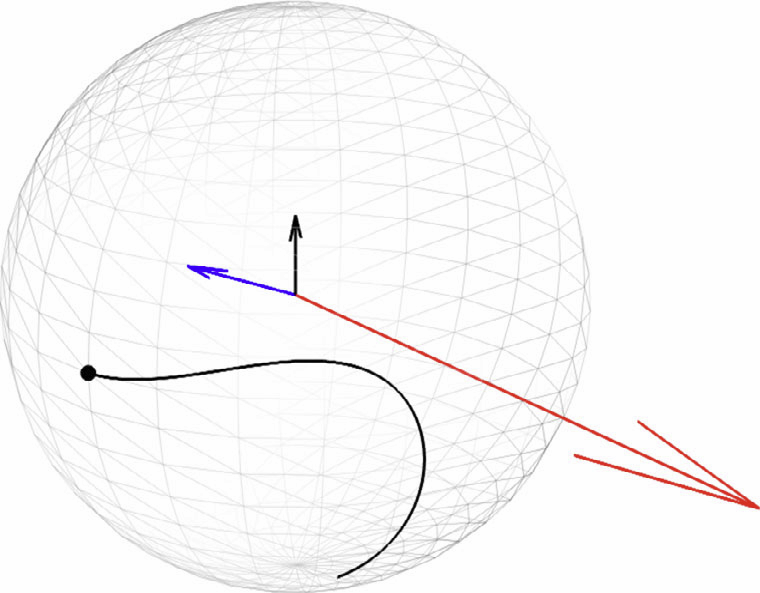
J. Hinkle, P.T. Fletcher, S. Joshi .
“Intrinsic Polynomials for Regression on Riemannian Manifolds,” In Journal of Mathematical Imaging and Vision, pp. 1-21. 2014.
T. Hollt, A. Magdy, P. Zhan, G. Chen, G. Gopalakrishnan, I. Hoteit, C.D. Hansen, M. Hadwiger.
“Ovis: A Framework for Visual Analysis of Ocean Forecast Ensembles,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. PP, No. 99, pp. 1. 2014.
DOI: 10.1109/TVCG.2014.2307892
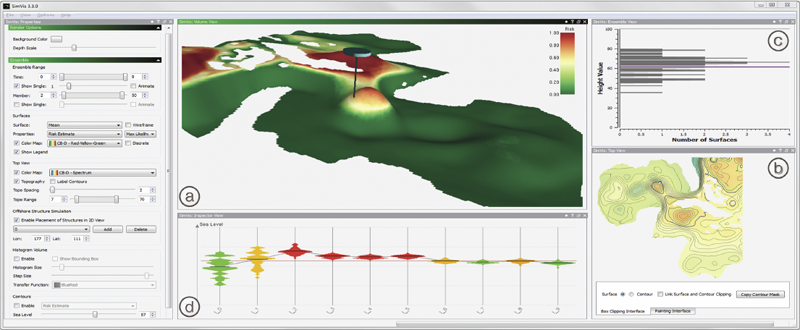
Keywords: Ensemble Visualization, Ocean Visualization, Ocean Forecast, Risk Estimation
J.B. Hoying, U. Utzinger, J.A. Weiss.
“Formation of microvascular networks: role of stromal interactions directing angiogenic growth,” In Microcirculation, Vol. 21, No. 4, pp. 278--289. May, 2014.
DOI: 10.1111/micc.12115
PubMed ID: 24447042
PubMed Central ID: PMC4032604
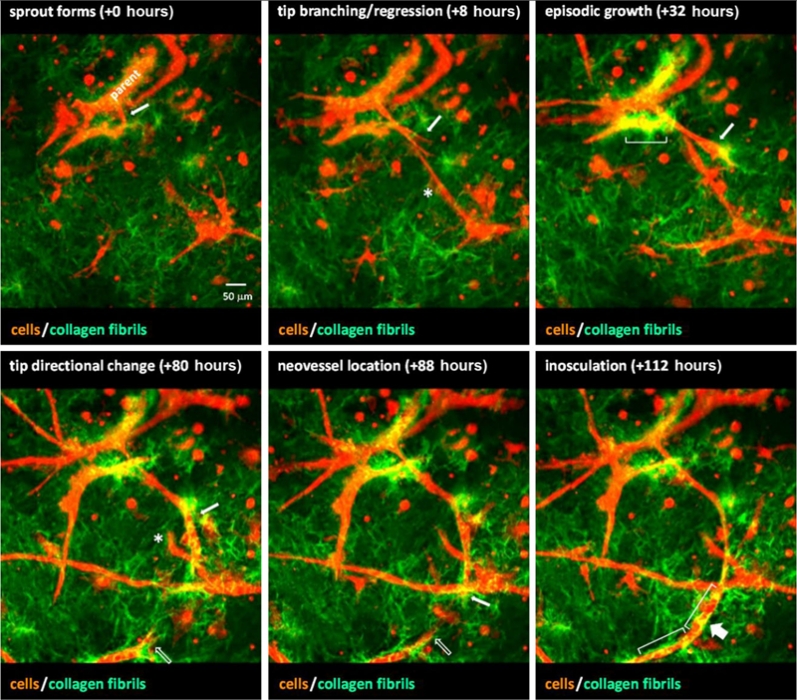
Keywords: angiogenesis, matrix, neovessel, remodeling, stroma
A. Humphrey, Q. Meng, M. Berzins, D. Caminha B.de Oliveira, Z. Rakamaric, G. Gopalakrishnan.
“Systematic Debugging Methods for Large-Scale HPC Computational Frameworks,” In Computing in Science Engineering, Vol. 16, No. 3, pp. 48--56. May, 2014.
ISSN: 1521-9615
DOI: 10.1109/MCSE.2014.11

Keywords: Computational Modeling and Frameworks, Parallel Programming, Reliability, Debugging Aids
Y. Joon Ahn, C. Hoffmann, P. Rosen.
“Geometric constraints on quadratic Bézier curves using minimal length and energy,” In Journal of Computational and Applied Mathematics, Vol. 255, pp. 887--897. 2014.
This paper derives expressions for the arc length and the bending energy of quadratic Bézier curves. The formulas are in terms of the control point coordinates. For fixed start and end points of the Bézier curve, the locus of the middle control point is analyzed for curves of fixed arc length or bending energy. In the case of arc length this locus is convex. For bending energy it is not. Given a line or a circle and fixed end points, the locus of the middle control point is determined for those curves that are tangent to a given line or circle. For line tangency, this locus is a parallel line. In the case of the circle, the locus can be classified into one of six major types. In some of these cases, the locus contains circular arcs. These results are then used to implement fast algorithms that construct quadratic Bézier curves tangent to a given line or circle, with given end points, that minimize bending energy or arc length.
A. Knoll, I. Wald, P. Navratil, A. Bowen, K. Reda, M. E. Papka, K. Gaither.
“RBF Volume Ray Casting on Multicore and Manycore CPUs,” In Computer Graphics Forum, Vol. 33, No. 3, Edited by H. Carr and P. Rheingans and H. Schumann, Wiley-Blackwell, pp. 71--80. June, 2014.
DOI: 10.1111/cgf.12363

S. Kumar, C. Christensen, P.-T. Bremer, E. Brugger, V. Pascucci, J. Schmidt, M. Berzins, H. Kolla, J. Chen, V. Vishwanath, P. Carns, R. Grout.
“Fast Multi-Resolution Reads of Massive Simulation Datasets,” In Proceedings of the International Supercomputing Conference ISC'14, Leipzig, Germany, June, 2014.
S. Kumar, J. Edwards, P.-T. Bremer, A. Knoll, C. Christensen, V. Vishwanath, P. Carns, J.A. Schmidt, V. Pascucci.
“Efficient I/O and storage of adaptive-resolution data,” In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, IEEE Press, pp. 413--423. 2014.
DOI: 10.1109/SC.2014.39

A.G. Landge, V. Pascucci, A. Gyulassy, J.C. Bennett, H. Kolla, J. Chen, P.-T. Bremer.
“In-situ feature extraction of large scale combustion simulations using segmented merge trees,” In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC 2014), New Orleans, Louisana, IEEE Press, Piscataway, NJ, USA pp. 1020--1031. 2014.
ISBN: 978-1-4799-5500-8
DOI: 10.1109/SC.2014.88
The ever increasing amount of data generated by scientific simulations coupled with system I/O constraints are fueling a need for in-situ analysis techniques. Of particular interest are approaches that produce reduced data representations while maintaining the ability to redefine, extract, and study features in a post-process to obtain scientific insights.
This paper presents two variants of in-situ feature extraction techniques using segmented merge trees, which encode a wide range of threshold based features. The first approach is a fast, low communication cost technique that generates an exact solution but has limited scalability. The second is a scalable, local approximation that nevertheless is guaranteed to correctly extract all features up to a predefined size. We demonstrate both variants using some of the largest combustion simulations available on leadership class supercomputers. Our approach allows state-of-the-art, feature-based analysis to be performed in-situ at significantly higher frequency than currently possible and with negligible impact on the overall simulation runtime.
J.D. Lewis, A.C. Evans, J.R. Pruett, K. Botteron, L. Zwaigenbaum, A. Estes, G. Gerig, L. Collins, P. Kostopoulos, R. McKinstry, S. Dager, S. Paterson, R. Schultz, M. Styner, H. Hazlett, J. Piven, IBIS network.
“Network inefficiencies in autism spectrum disorder at 24 months,” In Translational Psychiatry, Vol. 4, No. 5, Nature Publishing Group, pp. e388. May, 2014.
DOI: 10.1038/tp.2014.24
Autism Spectrum Disorder (ASD) is a developmental disorder defined by behavioural symptoms that emerge during the first years of life. Associated with these symptoms are differences in the structure of a wide array of brain regions, and in the connectivity between these regions. However, the use of cohorts with large age variability and participants past the generally recognized age of onset of the defining behaviours means that many of the reported abnormalities may be a result of cascade effects of developmentally earlier deviations. This study assessed differences in connectivity in ASD at the age at which the defining behaviours first become clear. The participants were 113 24-month-olds at high risk for ASD, 31 of whom were classified as ASD, and 23 typically developing 24-month-olds at low risk for ASD. Utilizing diffusion data to obtain measures of the length and strength of connections between anatomical regions, we performed an analysis of network efficiency. Our results showed significantly decreased local and global efficiency over temporal, parietal, and occipital lobes in high-risk infants classified as ASD, relative to both low- and high-risk infants not classified as ASD. The frontal lobes showed only a reduction in global efficiency in Broca's area. Additionally, these same regions showed an inverse relation between efficiency and symptom severity across the high-risk infants. The results suggest delay or deficits in infants with ASD in the optimization of both local and global aspects of network structure in regions involved in processing auditory and visual stimuli, language, and nonlinguistic social stimuli.
Keywords: autism, infant siblings, connectivity, network analysis, efficiency
A. Lex, N. Gehlenborg, H. Strobelt, R. Vuillemot,, H. Pfister.
“UpSet: Visualization of Intersecting Sets,” In IEEE Transactions on Visualization and Computer Graphics (InfoVis '14), Vol. 20, No. 12, pp. 1983--1992. 2014.
ISSN: 1077-2626
Understanding relationships between sets is an important analysis task that has received widespread attention in the visualization community. The major challenge in this context is the combinatorial explosion of the number of set intersections if the number of sets exceeds a trivial threshold. In this paper we introduce UpSet, a novel visualization technique for the quantitative analysis of sets, their intersections, and aggregates of intersections. UpSet is focused on creating task-driven aggregates, communicating the size and properties of aggregates and intersections, and a duality between the visualization of the elements in a dataset and their set membership. UpSet visualizes set intersections in a matrix layout and introduces aggregates based on groupings and queries. The matrix layout enables the effective representation of associated data, such as the number of elements in the aggregates and intersections, as well as additional summary statistics derived from subset or element attributes. Sorting according to various measures enables a task-driven analysis of relevant intersections and aggregates. The elements represented in the sets and their associated attributes are visualized in a separate view. Queries based on containment in specific intersections, aggregates or driven by attribute filters are propagated between both views. We also introduce several advanced visual encodings and interaction methods to overcome the problems of varying scales and to address scalability. UpSet is web-based and open source. We demonstrate its general utility in multiple use cases from various domains.
W. Liu, S.P. Awate, J.S. Anderson, P.T. Fletcher.
“A functional network estimation method of resting-state fMRI using a hierarchical Markov random field,” In NeuroImage, Vol. 100, pp. 520--534. 2014.
ISSN: 1053-8119
DOI: 10.1016/j.neuroimage.2014.06.001

Keywords: Resting-state functional MRI, Segmentation, Functional connectivity, Hierarchical Markov random field, Bayesian
Shusen Liu, Bei Wang, J.J. Thiagarajan, P.-T. Bremer, V. Pascucci.
“Visual Exploration of High-Dimensional Data: Subspace Analysis through Dynamic Projections,” SCI Technical Report, No. UUSCI-2014-003, SCI Institute, University of Utah, 2014.
Understanding high-dimensional data is rapidly becoming a central challenge in many areas of science and engineering. Most current techniques either rely on manifold learning based techniques which typically create a single embedding of the data or on subspace selection to find subsets of the original attributes that highlight the structure. However, the former creates a single, difficult-to-interpret view and assumes the data to be drawn from a single manifold, while the latter is limited to axis-aligned projections with restrictive viewing angles. Instead, we introduce ideas based on subspace clustering that can faithfully represent more complex data than the axis-aligned projections, yet do not assume the data to lie on a single manifold. In particular, subspace clustering assumes that the data can be represented by a union of low-dimensional subspaces, which can subsequently be used for analysis and visualization. In this paper, we introduce new techniques to reliably estimate both the intrinsic dimension and the linear basis of a mixture of subspaces extracted through subspace clustering. We show that the resulting bases represent the high-dimensional structures more reliably than traditional approaches. Subsequently, we use the bases to define different “viewpoints”, i.e., different projections onto pairs of basis vectors, from which to visualize the data. While more intuitive than non-linear projections, interpreting linear subspaces in terms of the original dimensions can still be challenging. To address this problem, we present new, animated transitions between different views to help the user navigate and explore the high-dimensional space. More specifically, we introduce the view transition graph which contains nodes for each subspace viewpoint and edges for potential transition between views. The transition graph enables users to explore both the structure within a subspace and the relations between different subspaces, for better understanding of the data. Using a number of case studies on well-know reference datasets, we demonstrate that the interactive exploration through such dynamic projections provides additional insights not readily available from existing tools.
Keywords: High-dimensional data, Subspace, Dynamic projection
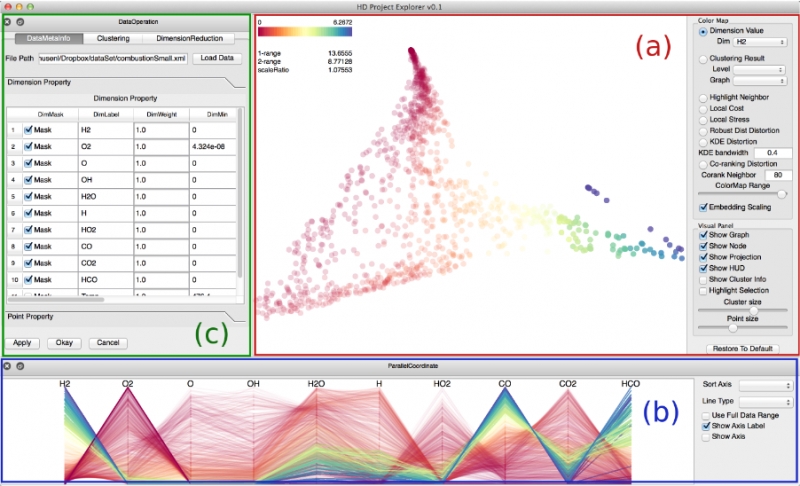
S. Liu, Bei Wang, P.-T. Bremer, V. Pascucci.
“Distortion-Guided Structure-Driven Interactive Exploration of High-Dimensional Data,” In Computer Graphics Forum, Vol. 33, No. 3, Wiley-Blackwell, pp. 101--110. June, 2014.
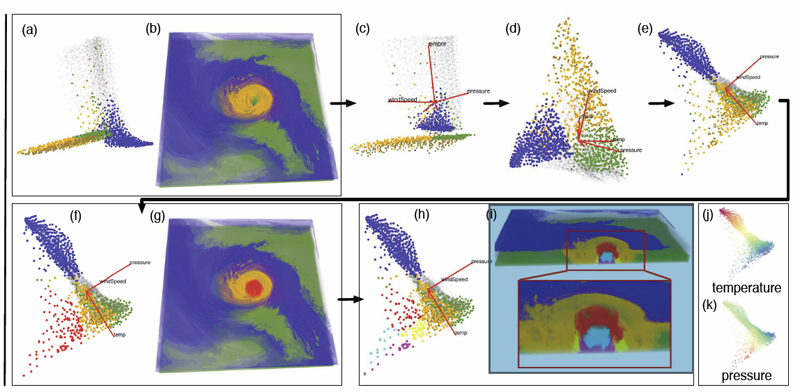
Shusen Liu, Bei Wang, J.J. Thiagarajan, P.-T. Bremer, V. Pascucci.
“Multivariate Volume Visualization through Dynamic Projections,” In Proceedings of the IEEE Symposium on Large Data Analysis and Visualization (LDAV), 2014.
Page 40 of 144
