SCI Publications
2012

S. Kole, N.P. Singh, R. King.
“Whole Brain Fractal Analysis of the Cerebral Cortex across the Adult Lifespan,” In Neurology, Meeting Abstracts I, Vol. 78, pp. P03.104. 2012.


D. Kopta, T. Ize, J. Spjut, E. Brunvand, A. Davis, A. Kensler.
“Fast, Effective BVH Updates for Animated Scenes,” In Proceedings of the Symposium on Interactive 3D Graphics and Games (I3D '12), pp. 197--204. 2012.
DOI: 10.1145/2159616.2159649
Bounding volume hierarchies (BVHs) are a popular acceleration structure choice for animated scenes rendered with ray tracing. This is due to the relative simplicity of refitting bounding volumes around moving geometry. However, the quality of such a refitted tree can degrade rapidly if objects in the scene deform or rearrange significantly as the animation progresses, resulting in dramatic increases in rendering times and a commensurate reduction in the frame rate. The BVH could be rebuilt on every frame, but this could take significant time. We present a method to efficiently extend refitting for animated scenes with tree rotations, a technique previously proposed for off-line improvement of BVH quality for static scenes. Tree rotations are local restructuring operations which can mitigate the effects that moving primitives have on BVH quality by rearranging nodes in the tree during each refit rather than triggering a full rebuild. The result is a fast, lightweight, incremental update algorithm that requires negligible memory, has minor update times, parallelizes easily, avoids significant degradation in tree quality or the need for rebuilding, and maintains fast rendering times. We show that our method approaches or exceeds the frame rates of other techniques and is consistently among the best options regardless of the animated scene.


S. Kumar, V. Vishwanath, P. Carns, J.A. Levine, R. Latham, G. Scorzelli, H. Kolla, R. Grout, R. Ross, M.E. Papka, J. Chen, V. Pascucci.
“Efficient data restructuring and aggregation for I/O acceleration in PIDX,” In Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, IEEE Computer Society Press, pp. 50:1--50:11. 2012.
ISBN: 978-1-4673-0804-5
Hierarchical, multiresolution data representations enable interactive analysis and visualization of large-scale simulations. One promising application of these techniques is to store high performance computing simulation output in a hierarchical Z (HZ) ordering that translates data from a Cartesian coordinate scheme to a one-dimensional array ordered by locality at different resolution levels. However, when the dimensions of the simulation data are not an even power of 2, parallel HZ ordering produces sparse memory and network access patterns that inhibit I/O performance. This work presents a new technique for parallel HZ ordering of simulation datasets that restructures simulation data into large (power of 2) blocks to facilitate efficient I/O aggregation. We perform both weak and strong scaling experiments using the S3D combustion application on both Cray-XE6 (65,536 cores) and IBM Blue Gene/P (131,072 cores) platforms. We demonstrate that data can be written in hierarchical, multiresolution format with performance competitive to that of native data-ordering methods.


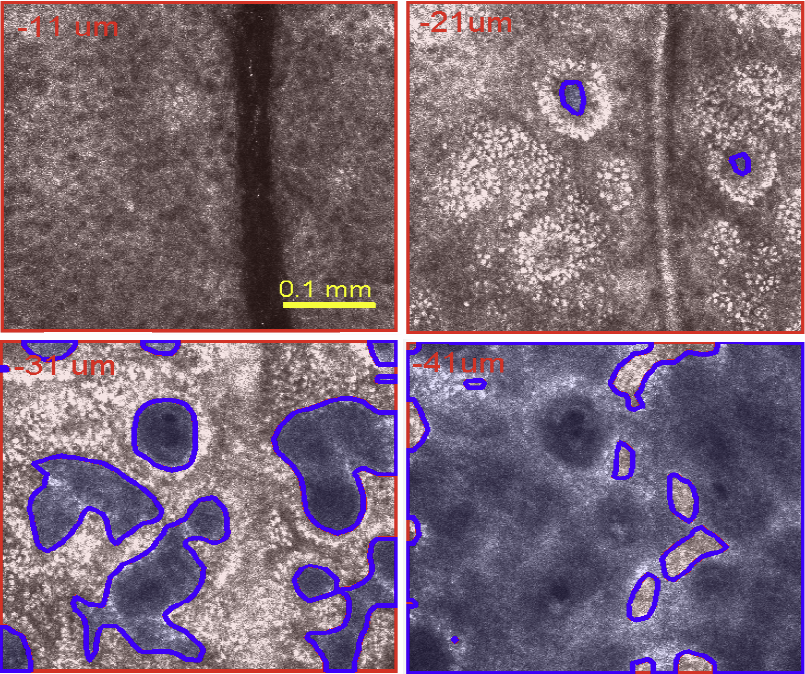
S. Kurugol, M. Rajadhyaksha, J.G. Dy, D.H. Brooks.
“Validation study of automated dermal/epidermal junction localization algorithm in reflectance confocal microscopy images of skin,” In Proceedings of SPIE Photonic Therapeutics and Diagnostics VIII, Vol. 8207, No. 1, pp. 820702-820711. 2012.
DOI: 10.1117/12.909227
PubMed ID: 24376908
PubMed Central ID: PMC3872972
Reflectance confocal microscopy (RCM) has seen increasing clinical application for noninvasive diagnosis of skin cancer. Identifying the location of the dermal-epidermal junction (DEJ) in the image stacks is key for effective clinical imaging. For example, one clinical imaging procedure acquires a dense stack of 0.5x0.5mm FOV images and then, after manual determination of DEJ depth, collects a 5x5mm mosaic at that depth for diagnosis. However, especially in lightly pigmented skin, RCM images have low contrast at the DEJ which makes repeatable, objective visual identification challenging. We have previously published proof of concept for an automated algorithm for DEJ detection in both highly- and lightly-pigmented skin types based on sequential feature segmentation and classification. In lightly-pigmented skin the change of skin texture with depth was detected by the algorithm and used to locate the DEJ. Here we report on further validation of our algorithm on a more extensive collection of 24 image stacks (15 fair skin, 9 dark skin). We compare algorithm performance against classification by three clinical experts. We also evaluate inter-expert consistency among the experts. The average correlation across experts was 0.81 for lightly pigmented skin, indicating the difficulty of the problem. The algorithm achieved epidermis/dermis misclassification rates smaller than 10% (based on 25x25 mm tiles) and average distance from the expert labeled boundaries of ~6.4 ?m for fair skin and ~5.3 ?m for dark skin, well within average cell size and less than 2x the instrument resolution in the optical axis.


A.G. Landge, J.A. Levine, A. Bhatele, K.E. Isaacs, T. Gamblin, S. Langer, M. Schulz, P.-T. Bremer, V. Pascucci.
“Visualizing Network Traffic to Understand the Performance of Massively Parallel Simulations,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 18, No. 12, IEEE, pp. 2467--2476. Dec, 2012.
DOI: 10.1109/TVCG.2012.286
The performance of massively parallel applications is often heavily impacted by the cost of communication among compute nodes. However, determining how to best use the network is a formidable task, made challenging by the ever increasing size and complexity of modern supercomputers. This paper applies visualization techniques to aid parallel application developers in understanding the network activity by enabling a detailed exploration of the flow of packets through the hardware interconnect. In order to visualize this large and complex data, we employ two linked views of the hardware network. The first is a 2D view, that represents the network structure as one of several simplified planar projections. This view is designed to allow a user to easily identify trends and patterns in the network traffic. The second is a 3D view that augments the 2D view by preserving the physical network topology and providing a context that is familiar to the application developers. Using the massively parallel multi-physics code pF3D as a case study, we demonstrate that our tool provides valuable insight that we use to explain and optimize pF3D’s performance on an IBM Blue Gene/P system.


B. Leavy, J. Clayton, O.E. Strack, R.M. Brannon, E. Strassburger.
“Edge on Impact Simulations and Experiments,” In Proceedings of the 12th Hypervelocity Impact Symposium, 2012.
In the quest to understand damage and failure of ceramics in ballistic events, simplified experiments have been developed to benchmark behavior. One such experiment is known as edge on impact (EOI). In this experiment, an impactor strikes the edge of a thin square plate, and damage and cracking that occur on the free surface are captured in real time with high speed photography. If the material of interest is transparent, additional information regarding damage and wave mechanics within the sample can be discerned. Polarizers can be used to monitor stress wave propagation, and photography can record internal damage. This information serves as an excellent benchmark for validation of ceramic and glass constitutive models implemented in dynamic simulation codes. In this report, recent progress towards predictive modeling of EOI is discussed. Time-dependent crack propagation and damage front evolution in silicon carbide and aluminum oxynitride ceramics are predicted using the Kayenta macroscopic constitutive model. Aspects regarding modeling material failure, variability, and volume scaling are noted. Mesoscale simulations of dynamic failure of anisotropic ceramic crystals facilitate determination of limit surfaces entering the macroscopic constitutive model, offsetting limited available experimental data.


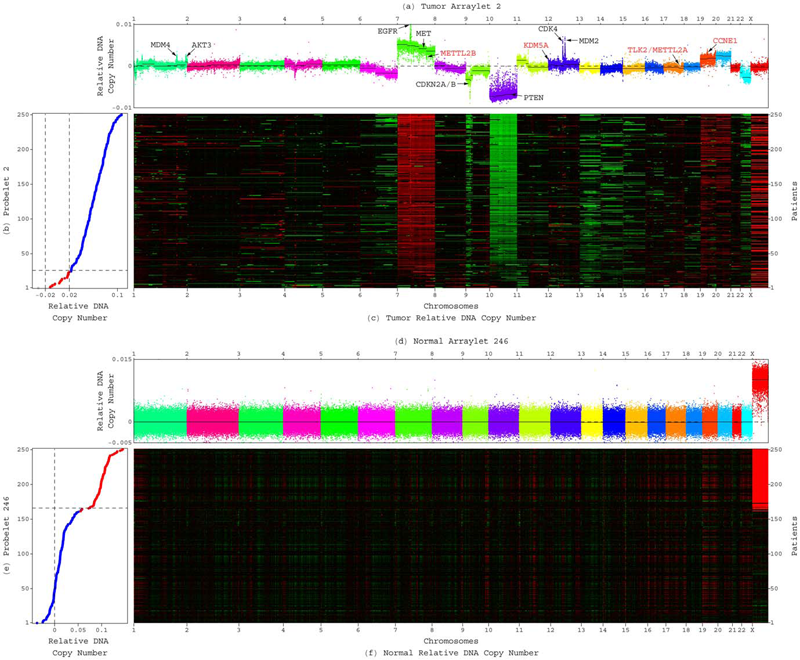
C.H. Lee, B.O. Alpert, P. Sankaranarayanan, O. Alter.
“GSVD Comparison of Patient-Matched Normal and Tumor aCGH Profiles Reveals Global Copy-Number Alterations Predicting Glioblastoma Multiforme Survival,” In PLoS ONE, Vol. 7, No. 1, Public Library of Science, pp. e30098. 2012.
DOI: 10.1371/journal.pone.0030098
Despite recent large-scale profiling efforts, the best prognostic predictor of glioblastoma multiforme (GBM) remains the patient's age at diagnosis. We describe a global pattern of tumor-exclusive co-occurring copy-number alterations (CNAs) that is correlated, possibly coordinated with GBM patients' survival and response to chemotherapy. The pattern is revealed by GSVD comparison of patient-matched but probe-independent GBM and normal aCGH datasets from The Cancer Genome Atlas (TCGA). We find that, first, the GSVD, formulated as a framework for comparatively modeling two composite datasets, removes from the pattern copy-number variations (CNVs) that occur in the normal human genome (e.g., female-specific X chromosome amplification) and experimental variations (e.g., in tissue batch, genomic center, hybridization date and scanner), without a-priori knowledge of these variations. Second, the pattern includes most known GBM-associated changes in chromosome numbers and focal CNAs, as well as several previously unreported CNAs in greater than 3\% of the patients. These include the biochemically putative drug target, cell cycle-regulated serine/threonine kinase-encoding TLK2, the cyclin E1-encoding CCNE1, and the Rb-binding histone demethylase-encoding KDM5A. Third, the pattern provides a better prognostic predictor than the chromosome numbers or any one focal CNA that it identifies, suggesting that the GBM survival phenotype is an outcome of its global genotype. The pattern is independent of age, and combined with age, makes a better predictor than age alone. GSVD comparison of matched profiles of a larger set of TCGA patients, inclusive of the initial set, confirms the global pattern. GSVD classification of the GBM profiles of an independent set of patients validates the prognostic contribution of the pattern.


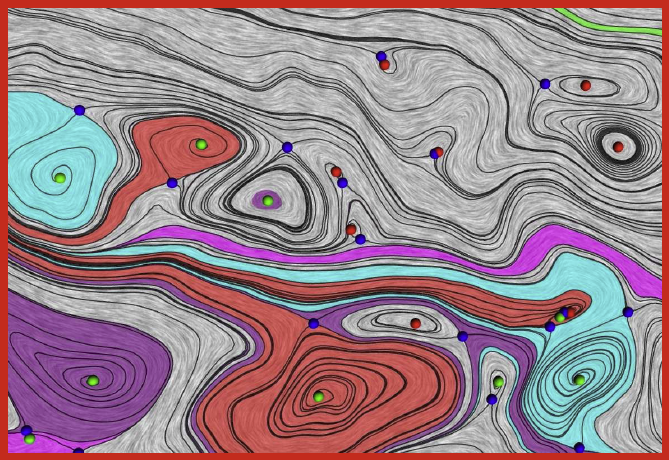
J.A. Levine, S. Jadhav, H. Bhatia, V. Pascucci, P.-T. Bremer.
“A Quantized Boundary Representation of 2D Flows,” In Computer Graphics Forum, Vol. 31, No. 3 Pt. 1, pp. 945--954. June, 2012.
DOI: 10.1111/j.1467-8659.2012.03087.x
Analysis and visualization of complex vector fields remain major challenges when studying large scale simulation of physical phenomena. The primary reason is the gap between the concepts of smooth vector field theory and their computational realization. In practice, researchers must choose between either numerical techniques, with limited or no guarantees on how they preserve fundamental invariants, or discrete techniques which limit the precision at which the vector field can be represented. We propose a new representation of vector fields that combines the advantages of both approaches. In particular, we represent a subset of possible streamlines by storing their paths as they traverse the edges of a triangulation. Using only a finite set of streamlines creates a fully discrete version of a vector field that nevertheless approximates the smooth flow up to a user controlled error bound. The discrete nature of our representation enables us to directly compute and classify analogues of critical points, closed orbits, and other common topological structures. Further, by varying the number of divisions (quantizations) used per edge, we vary the resolution used to represent the field, allowing for controlled precision. This representation is compact in memory and supports standard vector field operations.


A. Lex, M. Streit, H. Schulz, C. Partl, D. Schmalstieg, P.. Park, N. Gehlenborg.
“StratomeX: Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization ,” In Computer Graphics Forum (EuroVis '12), Vol. 31, No. 3, pp. 1175--1184. 2012.
ISSN: 0167-7055
DOI: 10.1111/j.1467-8659.2012.03110.x
dentification and characterization of cancer subtypes are important areas of research that are based on the integrated analysis of multiple heterogeneous genomics datasets. Since there are no tools supporting this process, much of this work is done using ad-hoc scripts and static plots, which is inefficient and limits visual exploration of the data. To address this, we have developed StratomeX, an integrative visualization tool that allows investigators to explore the relationships of candidate subtypes across multiple genomic data types such as gene expression, DNA methylation, or copy number data. StratomeX represents datasets as columns and subtypes as bricks in these columns. Ribbons between the columns connect bricks to show subtype relationships across datasets. Drill-down features enable detailed exploration. StratomeX provides insights into the functional and clinical implications of candidate subtypes by employing small multiples, which allow investigators to assess the effect of subtypes on molecular pathways or outcomes such as patient survival. As the configuration of viewing parameters in such a multi-dataset, multi-view scenario is complex, we propose a meta visualization and configuration interface for dataset dependencies and data-view relationships. StratomeX is developed in close collaboration with domain experts. We describe case studies that illustrate how investigators used the tool to explore subtypes in large datasets and demonstrate how they efficiently replicated findings from the literature and gained new insights into the data.


J. Li, D. Xiu.
“Computation of Failure Probability Subject to Epistemic Uncertainty,” In SIAM Journal on Scientific Computing, Vol. 34, No. 6, pp. A2946--A2964. 2012.
DOI: 10.1137/120864155
Computing failure probability is a fundamental task in many important practical problems. The computation, its numerical challenges aside, naturally requires knowledge of the probability distribution of the underlying random inputs. On the other hand, for many complex systems it is often not possible to have complete information about the probability distributions. In such cases the uncertainty is often referred to as epistemic uncertainty, and straightforward computation of the failure probability is not available. In this paper we develop a method to estimate both the upper bound and the lower bound of the failure probability subject to epistemic uncertainty. The bounds are rigorously derived using the variational formulas for relative entropy. We examine in detail the properties of the bounds and present numerical algorithms to efficiently compute them.
Keywords: failure probability, uncertainty quantification, epistemic uncertainty, relative entropy


T. Liu, E. Jurrus, M. Seyedhosseini, M. Ellisman, T. Tasdizen.
“Watershed Merge Tree Classification for Electron Microscopy Image Segmentation,” In Proceedings of the 21st International Conference on Pattern Recognition (ICPR), pp. 133--137. 2012.
Automated segmentation of electron microscopy (EM) images is a challenging problem. In this paper, we present a novel method that utilizes a hierarchical structure and boundary classification for 2D neuron segmentation. With a membrane detection probability map, a watershed merge tree is built for the representation of hierarchical region merging from the watershed algorithm. A boundary classifier is learned with non-local image features to predict each potential merge in the tree, upon which merge decisions are made with consistency constraints to acquire the final segmentation. Independent of classifiers and decision strategies, our approach proposes a general framework for efficient hierarchical segmentation with statistical learning. We demonstrate that our method leads to a substantial improvement in segmentation accuracy.


S. Liu, J.A. Levine, P.-T. Bremer, V. Pascucci.
“Gaussian Mixture Model Based Volume Visualization,” In Proceedings of the IEEE Large-Scale Data Analysis and Visualization Symposium 2012, Note: Received Best Paper Award, pp. 73--77. 2012.
DOI: 10.1109/LDAV.2012.6378978
Representing uncertainty when creating visualizations is becoming more indispensable to understand and analyze scientific data. Uncertainty may come from different sources, such as, ensembles of experiments or unavoidable information loss when performing data reduction. One natural model to represent uncertainty is to assume that each position in space instead of a single value may take on a distribution of values. In this paper we present a new volume rendering method using per voxel Gaussian mixture models (GMMs) as the input data representation. GMMs are an elegant and compact way to drastically reduce the amount of data stored while still enabling realtime data access and rendering on the GPU. Our renderer offers efficient sampling of the data distribution, generating renderings of the data that flicker at each frame to indicate high variance. We can accumulate samples as well to generate still frames of the data, which preserve additional details in the data as compared to either traditional scalar indicators (such as a mean or a single nearest neighbor down sample) or to fitting the data with only a single Gaussian per voxel. We demonstrate the effectiveness of our method using ensembles of climate simulations and MRI scans as well as the down sampling of large scalar fields as examples.
Keywords: Uncertainty Visualization, Volume Rendering, Gaussian Mixture Model, Ensemble Visualization


W. Liu, S. Awate, P.T. Fletcher.
“Group Analysis of Resting-State fMRI by Hierarchical Markov Random Fields,” In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2012, Lecture Notes in Computer Science (LNCS), Vol. 7512, pp. 189--196. 2012.
ISBN: 978-3-642-33453-5
DOI: 10.1007/978-3-642-33454-2_24
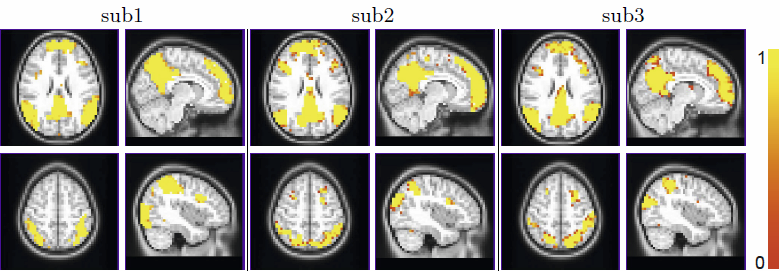
Identifying functional networks from resting-state functional MRI is a challenging task, especially for multiple subjects. Most current studies estimate the networks in a sequential approach, i.e., they identify each individual subject's network independently to other subjects, and then estimate the group network from the subjects networks. This one-way flow of information prevents one subject's network estimation benefiting from other subjects. We propose a hierarchical Markov Random Field model, which takes into account both the within-subject spatial coherence and between-subject consistency of the network label map. Both population and subject network maps are estimated simultaneously using a Gibbs sampling approach in a Monte Carlo Expectation Maximization framework. We compare our approach to two alternative groupwise fMRI clustering methods, based on K-means and Normalized Cuts, using both synthetic and real fMRI data.We show that our method is able to estimate more consistent subject label maps, as well as a stable group label map.


Y. Livnat, T.-M. Rhyne, M. Samore.
“Epinome: A Visual-Analytics Workbench for Epidemiology Data,” In IEEE Computer Graphics and Applications, Vol. 32, No. 2, pp. 89--95. 2012.
ISSN: 0272-1716
DOI: 10.1109/MCG.2012.31
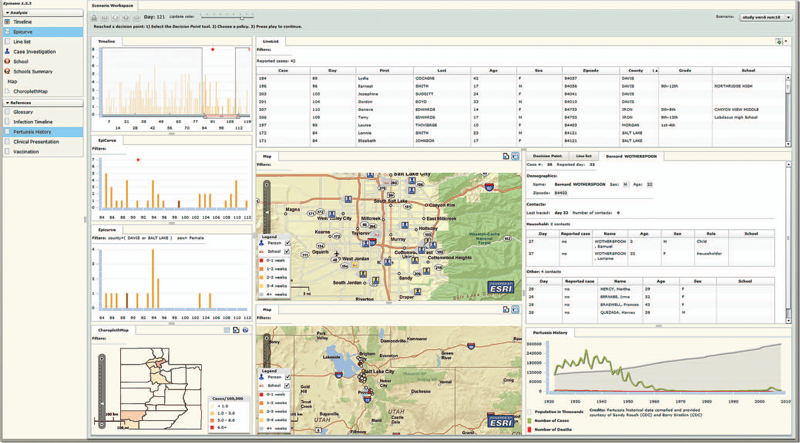
Effective detection of and response to infectious disease outbreaks depend on the ability to capture and analyze information and on how public health officials respond to this information. Researchers have developed various surveillance systems to automate data collection, analysis, and alert generation, yet the massive amount of collected data often leads to information overload. To improve decision-making in outbreak detection and response, it's important to understand how outbreak investigators seek relevant information. Studying their information-search strategies can provide insight into their cognitive biases and heuristics. Identifying the presence of such biases will enable the development of tools that counter balance them and help users develop alternative scenarios.
We implemented a large-scale high-fidelity simulation of scripted infectious-disease outbreaks to help us study public health practitioners' information- search strategies. We also developed Epinome, an integrated visual-analytics investigation system. Epinome caters to users' needs by providing a variety of investigation tools. It facilitates user studies by recording which tools they used, when, and how. (See the video demonstration of Epinome at www.sci.utah.edu/gallery2/v/ software/epinome.) Epinome provides a dynamic environment that seamlessly evolves and adapts to user tasks and needs. It introduces four userinteraction paradigms in public health:
• an evolving visual display,
• seamless integration between disparate views,
• loosely coordinated multiple views, and
• direct interaction with data items.
Using Epinome, users can replay simulation scenarios, investigate an unfolding outbreak using a variety of visualization tools, and steer the simulation by implementing different public health policies at predefined decision points. Epinome records user actions, such as tool selection, interactions with each tool, and policy changes, and stores them in a database for postanalysis. A psychology team can then use that information to study users' search strategies.


H. Lu, M. Berzins, C.E. Goodyer, P.K. Jimack.
“Adaptive High-Order Discontinuous Galerkin Solution of Elastohydrodynamic Lubrication Point Contact Problems,” In Advances in Engineering Software, Vol. 45, No. 1, pp. 313--324. 2012.
DOI: 10.1016/j.advengsoft.2011.10.006
This paper describes an adaptive implementation of a high order Discontinuous Galerkin (DG) method for the solution of elastohydrodynamic lubrication (EHL) point contact problems. These problems arise when modelling the thin lubricating film between contacts which are under sufficiently high pressure that the elastic deformation of the contacting elements cannot be neglected. The governing equations are highly nonlinear and include a second order partial differential equation that is derived via the thin-film approximation. Furthermore, the problem features a free boundary, which models where cavitation occurs, and this is automatically captured as part of the solution process. The need for spatial adaptivity stems from the highly variable length scales that are present in typical solutions. Results are presented which demonstrate both the effectiveness and the limitations of the proposed adaptive algorithm.
Keywords: Elastohydrodynamic lubrication, Discontinuous Galerkin, High polynomial degree, h-adaptivity, Nonlinear systems


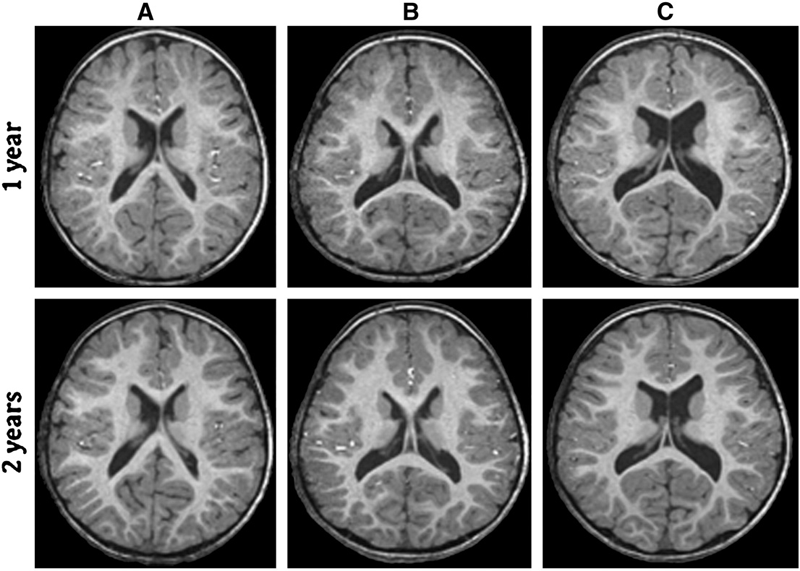
A.E. Lyall, S. Woolson, H.M. Wolf, B.D. Goldman, J.S. Reznick, R.M. Hamer, W. Lin, M. Styner, G. Gerig, J.H. Gilmore.
“Prenatal isolated mild ventriculomegaly is associated with persistent ventricle enlargement at ages 1 and 2,” In Early Human Development, Elsevier, pp. (in press). 2012.
Background: Enlargement of the lateral ventricles is thought to originate from abnormal prenatal brain development and is associated with neurodevelopmental disorders. Fetal isolated mild ventriculomegaly (MVM) is associated with the enlargement of lateral ventricle volumes in the neonatal period and developmental delays in early childhood. However, little is known about postnatal brain development in these children.
Methods: Twenty-eight children with fetal isolated MVM and 56 matched controls were followed at ages 1 and 2 years with structural imaging on a 3T Siemens scanner and assessment of cognitive development with the Mullen Scales of Early Learning. Lateral ventricle, total gray and white matter volumes, and Mullen cognitive composite scores and subscale scores were compared between groups.
Results: Compared to controls, children with prenatal isolated MVM had significantly larger lateral ventricle volumes at ages 1 and 2 years. Lateral ventricle volume at 1 and 2 years of age was significantly correlated with prenatal ventricle size. Enlargement of the lateral ventricles was associated with increased intracranial volumes and increased gray and white matter volumes. Children with MVM had Mullen composite scores similar to controls, although there was evidence of delay in fine motor and expressive language skills.
Conclusions: Children with prenatal MVM have persistent enlargement of the lateral ventricles through the age of 2 years; this enlargement is associated with increased gray and white matter volumes and some evidence of delay in fine motor and expressive language development. Further study is needed to determine if enlarged lateral ventricles are associated with increased risk for neurodevelopmental disorders.


S.A. Maas, B.J. Ellis, G.A. Ateshian, J.A. Weiss.
“FEBio: Finite elements for biomechanics,” In Journal of Biomechanical Engineering, Vol. 134, No. 1, pp. 011005. 2012.
DOI: 10.1115/1.4005694
PubMed ID: 22482660
In the field of computational biomechanics, investigators have primarily used commercial software that is neither geared toward biological applications nor sufficiently flexible to follow the latest developments in the field. This lack of a tailored software environment has hampered research progress, as well as dissemination of models and results. To address these issues, we developed the FEBio software suite (http://febio.org/), a nonlinear implicit finite element (FE) framework, designed specifically for analysis in computational solid biomechanics. This paper provides an overview of the theoretical basis of FEBio and its main features. FEBio offers modeling scenarios, constitutive models, and boundary conditions, which are relevant to numerous applications in biomechanics. The open-source FEBio software is written in C++, with particular attention to scalar and parallel performance on modern computer architectures. Software verification is a large part of the development and maintenance of FEBio, and to demonstrate the general approach, the description and results of several problems from the FEBio Verification Suite are presented and compared to analytical solutions or results from other established and verified FE codes. An additional simulation is described that illustrates the application of FEBio to a research problem in biomechanics. Together with the pre- and postprocessing software PREVIEW and POSTVIEW, FEBio provides a tailored solution for research and development in computational biomechanics.


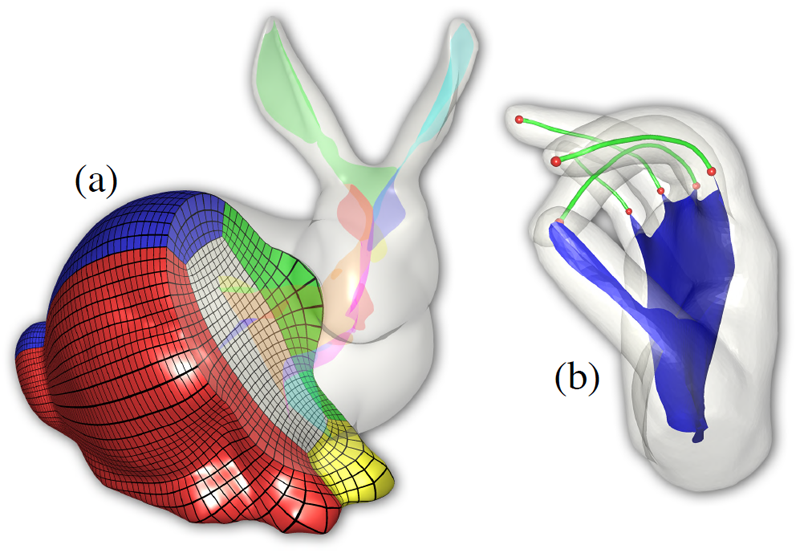
T. Martin, G. Chen, S. Musuvathy, E. Cohen, C.D. Hansen.
“Generalized Swept Mid-structure for Polygonal Models,” In Computer Graphics Forum, Vol. 31, No. 2 part 4, Wiley-Blackwell, pp. 805--814. May, 2012.
DOI: 10.1111/j.1467-8659.2012.03061.x
We introduce a novel mid-structure called the generalized swept mid-structure (GSM) of a closed polygonal shape, and a framework to compute it. The GSM contains both curve and surface elements and has consistent sheet-by-sheet topology, versus triangle-by-triangle topology produced by other mid-structure methods. To obtain this structure, a harmonic function, defined on the volume that is enclosed by the surface, is used to decompose the volume into a set of slices. A technique for computing the 1D mid-structures of these slices is introduced. The mid-structures of adjacent slices are then iteratively matched through a boundary similarity computation and triangulated to form the GSM. This structure respects the topology of the input surface model is a hybrid mid-structure representation. The construction and topology of the GSM allows for local and global simplification, used in further applications such as parameterization, volumetric mesh generation and medical applications.
Keywords: scidac, kaust


K.S. McDowell, F. Vadakkumpadan, R. Blake, J. Blauer, G. Plank, R.S. MacLeod, N.A. Trayanova.
“Methodology for patient-specific modeling of atrial fibrosis as a substrate for atrial fibrillation,” In Journal of Electrocardiology, Vol. 45, No. 6, pp. 640--645. 2012.
DOI: 10.1016/j.jelectrocard.2012.08.005
PubMed ID: 22999492
PubMed Central ID: PMC3515859
Personalized computational cardiac models are emerging as an important tool for studying cardiac arrhythmia mechanisms, and have the potential to become powerful instruments for guiding clinical anti-arrhythmia therapy. In this article, we present the methodology for constructing a patient-specific model of atrial fibrosis as a substrate for atrial fibrillation. The model is constructed from high-resolution late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) images acquired in vivo from a patient suffering from persistent atrial fibrillation, accurately capturing both the patient's atrial geometry and the distribution of the fibrotic regions in the atria. Atrial fiber orientation is estimated using a novel image-based method, and fibrosis is represented in the patient-specific fibrotic regions as incorporating collagenous septa, gap junction remodeling, and myofibroblast proliferation. A proof-of-concept simulation result of reentrant circuits underlying atrial fibrillation in the model of the patient's fibrotic atrium is presented to demonstrate the completion of methodology development.
Keywords: Patient-specific modeling, Computational model, Atrial fibrillation, Atrial fibrosis


Q. Meng, M. Berzins.
“Scalable Large-scale Fluid-structure Interaction Solvers in the Uintah Framework via Hybrid Task-based Parallelism Algorithms,” SCI Technical Report, No. UUSCI-2012-004, SCI Institute, University of Utah, 2012.
Uintah is a software framework that provides an environment for solving fluid-structure interaction problems on structured adaptive grids on large-scale science and engineering problems involving the solution of partial differential equations. Uintah uses a combination of fluid-flow solvers and particle-based methods for solids, together with adaptive meshing and a novel asynchronous task-based approach with fully automated load balancing. When applying Uintah to fluid-structure interaction problems with mesh refinement, the combination of adaptive meshing and the movement of structures through space present a formidable challenge in terms of achieving scalability on large-scale parallel computers. With core counts per socket continuing to grow along with the prospect of less memory per core, adopting a model that uses MPI to communicate between nodes and a shared memory model on-node is one approach to achieve scalability at full machine capacity on current and emerging large-scale systems. For this approach to be successful, it is necessary to design data-structures that large numbers of cores can simultaneously access without contention. These data structures and algorithms must also be designed to avoid the overhead involved with locks and other synchronization primitives when running on large number of cores per node, as contention for acquiring locks quickly becomes untenable. This scalability challenge is addressed here for Uintah, by the development of new hybrid runtime and scheduling algorithms combined with novel lockfree data structures, making it possible for Uintah to achieve excellent scalability for a challenging fluid-structure problem with mesh refinement on as many as 260K cores.
Keywords: uintah, csafe