SCI Publications
2013


S.P. Reese, C.J. Underwood, J.A. Weiss.
“Effects of decorin proteoglycan on fibrillogenesis, ultrastructure, and mechanics of type I collagen gels,” In Matrix Biology, pp. (in press). 2013.
DOI: 10.1016/j.matbio.2013.04.004
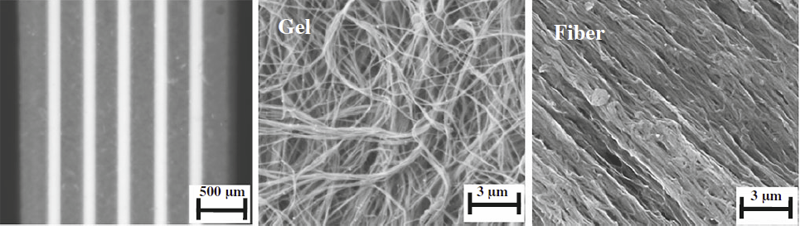
The proteoglycan decorin is known to affect both the fibrillogenesis and the resulting ultrastructure of in vitro polymerized collagen gels. However, little is known about its effects on mechanical properties. In this study, 3D collagen gels were polymerized into tensile test specimens in the presence of decorin proteoglycan, decorin core protein, or dermatan sulfate (DS). Collagen fibrillogenesis, ultrastructure, and mechanical properties were then quantified using a turbidity assay, 2 forms of microscopy (SEM and confocal), and tensile testing. The presence of decorin proteoglycan or core protein decreased the rate and ultimate turbidity during fibrillogenesis and decreased the number of fibril aggregates (fibers) compared to control gels. The addition of decorin and core protein increased the linear modulus by a factor of 2 compared to controls, while the addition of DS reduced the linear modulus by a factor of 3. Adding decorin after fibrillogenesis had no effect, suggesting that decorin must be present during fibrillogenesis to increase the mechanical properties of the resulting gels. These results show that the inclusion of decorin proteoglycan during fibrillogenesis of type I collagen increases the modulus and tensile strength of resulting collagen gels. The increase in mechanical properties when polymerization occurs in the presence of the decorin proteoglycan is due to a reduction in the aggregation of fibrils into larger order structures such as fibers and fiber bundles.


S.P. Reese, B.J. Ellis, J.A. Weiss.
“Micromechanical model of a surrogate for collagenous soft tissues: development, validation, and analysis of mesoscale size effects,” In Biomechanics and Modeling in Mechanobiology, pp. (in press). 2013.
DOI: 10.1007/s10237-013-0475-2
Aligned, collagenous tissues such as tendons and ligaments are composed primarily of water and type I collagen, organized hierarchically into nanoscale fibrils, microscale fibers and mesoscale fascicles. Force transfer across scales is complex and poorly understood. Since innervation, the vasculature, damage mechanisms and mechanotransduction occur at the microscale and mesoscale, understanding multiscale interactions is of high importance. This study used a physical model in combination with a computational model to isolate and examine the mechanisms of force transfer between scales. A collagen-based surrogate served as the physical model. The surrogate consisted of extruded collagen fibers embedded within a collagen gel matrix. A micromechanical finite element model of the surrogate was validated using tensile test data that were recorded using a custom tensile testing device mounted on a confocal microscope. Results demonstrated that the experimentally measured macroscale strain was not representative of the microscale strain, which was highly inhomogeneous. The micromechanical model, in combination with a macroscopic continuum model, revealed that the microscale inhomogeneity resulted from size effects in the presence of a constrained boundary. A sensitivity study indicated that significant scale effects would be present over a range of physiologically relevant inter-fiber spacing values and matrix material properties. The results indicate that the traditional continuum assumption is not valid for describing the macroscale behavior of the surrogate and that boundary-induced size effects are present.


P. Rosen, B. Burton, K. Potter, C.R. Johnson.
“Visualization for understanding uncertainty in the simulation of myocardial ischemia,” In Proceedings of the 2013 Workshop on Visualization in Medicine and Life Sciences, 2013.
We have created the Myocardial Uncertainty Viewer (muView) tool for exploring data stemming from the forward simulation of cardiac ischemia. The simulation uses a collection of conductivity values to understand how ischemic regions effect the undamaged anisotropic heart tissue. The data resulting from the simulation is multivalued and volumetric and thus, for every data point, we have a collection of samples describing cardiac electrical properties. muView combines a suite of visual analysis methods to explore the area surrounding the ischemic zone and identify how perturbations of variables changes the propagation of their effects.


P. Rosen.
“A Visual Approach to Investigating Shared and Global Memory Behavior of CUDA Kernels,” In Computer Graphics Forum, Vol. 32, No. 3, Wiley-Blackwell, pp. 161--170. June, 2013.
DOI: 10.1111/cgf.12103
We present an approach to investigate the memory behavior of a parallel kernel executing on thousands of threads simultaneously within the CUDA architecture. Our top-down approach allows for quickly identifying any significant differences between the execution of the many blocks and warps. As interesting warps are identified, we allow further investigation of memory behavior by visualizing the shared memory bank conflicts and global memory coalescence, first with an overview of a single warp with many operations and, subsequently, with a detailed view of a single warp and a single operation. We demonstrate the strength of our approach in the context of a parallel matrix transpose kernel and a parallel 1D Haar Wavelet transform kernel.


A. Rungta, B. Summa, D. Demir, P.-T. Bremer, V. Pascucci.
“ManyVis: Multiple Applications in an Integrated Visualization Environment,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 19, No. 12, pp. 2878--2885. December, 2013.
As the visualization field matures, an increasing number of general toolkits are developed to cover a broad range of applications. However, no general tool can incorporate the latest capabilities for all possible applications, nor can the user interfaces and workflows be easily adjusted to accommodate all user communities. As a result, users will often chose either substandard solutions presented in familiar, customized tools or assemble a patchwork of individual applications glued through ad-hoc scripts and extensive, manual intervention. Instead, we need the ability to easily and rapidly assemble the best-in-task tools into custom interfaces and workflows to optimally serve any given application community. Unfortunately, creating such meta-applications at the API or SDK level is difficult, time consuming, and often infeasible due to the sheer variety of data models, design philosophies, limits in functionality, and the use of closed commercial systems. In this paper, we present the ManyVis framework which enables custom solutions to be built both rapidly and simply by allowing coordination and communication across existing unrelated applications. ManyVis allows users to combine software tools with complementary characteristics into one virtual application driven by a single, custom-designed interface.


N. Sadeghi, M.W. Prastawa, P.T. Fletcher, C. Vachet, Bo Wang, J.H. Gilmore, G. Gerig.
“Multivariate Modeling of Longitudinal MRI in Early Brain Development with Confidence Measures,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 1400--1403. 2013.
DOI: 10.1109/ISBI.2013.6556795
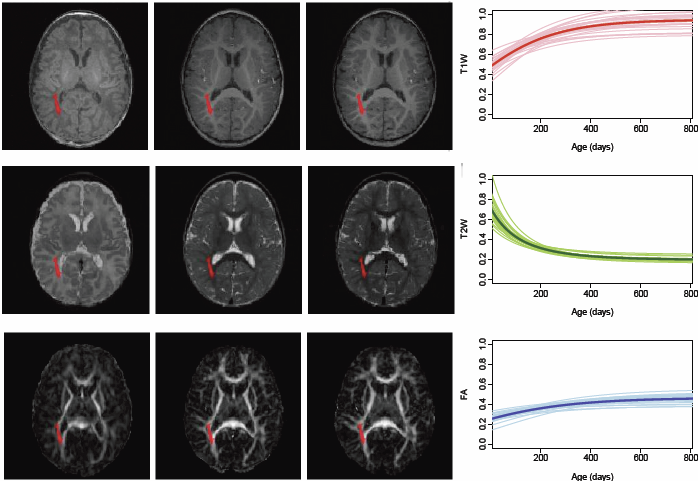
The human brain undergoes rapid organization and structuring early in life. Longitudinal imaging enables the study of these changes over a developmental period within individuals through estimation of population growth trajectory and its variability. In this paper, we focus on maturation of white and gray matter as is depicted in structural and diffusion MRI of healthy subjects with repeated scans. We provide a framework for joint analysis of both structural MRI and DTI (Diffusion Tensor Imaging) using multivariate nonlinear mixed effect modeling of temporal changes. Our framework constructs normative growth models for all the modalities that take into account the correlation among the modalities and individuals, along with estimation of the variability of the population trends. We apply our method to study early brain development, and to our knowledge this is the first multimodel longitudinal modeling of diffusion and signal intensity changes for this growth stage. Results show the potential of our framework to study growth trajectories, as well as neurodevelopmental disorders through comparison against the constructed normative models of multimodal 4D MRI.


N. Sadeghi, M.W. Prastawa, P.T. Fletcher, J. Wolff, J.H. Gilmore, G. Gerig.
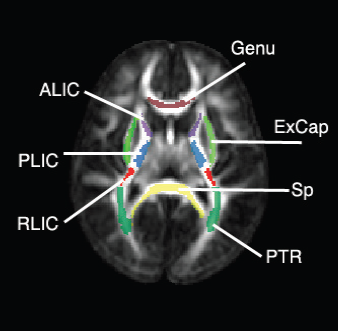
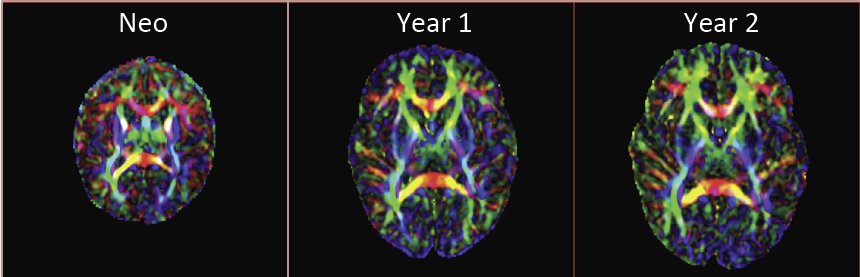
“Regional characterization of longitudinal DT-MRI to study white matter maturation of the early developing brain,” In NeuroImage, Vol. 68, pp. 236--247. March, 2013.
DOI: 10.1016/j.neuroimage.2012.11.040
PubMed ID: 23235270
The human brain undergoes rapid and dynamic development early in life. Assessment of brain growth patterns relevant to neurological disorders and disease requires a normative population model of growth and variability in order to evaluate deviation from typical development. In this paper, we focus on maturation of brain white matter as shown in diffusion tensor MRI (DT-MRI), measured by fractional anisotropy (FA), mean diffusivity (MD), as well as axial and radial diffusivities (AD, RD). We present a novel methodology to model temporal changes of white matter diffusion from longitudinal DT-MRI data taken at discrete time points. Our proposed framework combines nonlinear modeling of trajectories of individual subjects, population analysis, and testing for regional differences in growth pattern. We first perform deformable mapping of longitudinal DT-MRI of healthy infants imaged at birth, 1 year, and 2 years of age, into a common unbiased atlas. An existing template of labeled white matter regions is registered to this atlas to define anatomical regions of interest. Diffusivity properties of these regions, presented over time, serve as input to the longitudinal characterization of changes. We use non-linear mixed effect (NLME) modeling where temporal change is described by the Gompertz function. The Gompertz growth function uses intuitive parameters related to delay, rate of change, and expected asymptotic value; all descriptive measures which can answer clinical questions related to quantitative analysis of growth patterns. Results suggest that our proposed framework provides descriptive and quantitative information on growth trajectories that can be interpreted by clinicians using natural language terms that describe growth. Statistical analysis of regional differences between anatomical regions which are known to mature differently demonstrates the potential of the proposed method for quantitative assessment of brain growth and differences thereof. This will eventually lead to a prediction of white matter diffusion properties and associated cognitive development at later stages given imaging data at early stages.



N. Sadeghi, C. Vachet, M. Prastawa, J. Korenberg, G. Gerig.
“Analysis of Diffusion Tensor Imaging for Subjects with Down Syndrome,” In Proceedings of the 19th Annual Meeting of the Organization for Human Brain Mapping OHBM, pp. (in print). 2013.
Down syndrome (DS) is the most common chromosome abnormality in humans. It is typically associated with delayed cognitive development and physical growth. DS is also associated with Alzheimer-like dementia [1]. In this study we analyze the white matter integrity of individuals with DS compared to control as is reflected in the diffusion parameters derived from Diffusion Tensor Imaging. DTI provides relevant information about the underlying tissue, which correlates with cognitive function [2]. We present a cross-sectional analysis of white matter tracts of subjects with DS compared to control.


N. Sadeghi.
“Modeling and Analysis of Longitudinal Multimodal Magnetic Resonance Imaging: Application to Early Brain Development,” Note: Ph.D. Thesis, Department of Bioengineering, University of Utah, December, 2013.
Many mental illnesses are thought to have their origins in early stages of development, encouraging increased research efforts related to early neurodevelopment. Magnetic resonance imaging (MRI) has provided us with an unprecedented view of the brain in vivo. More recently, diffusion tensor imaging (DTI/DT-MRI), a magnetic resonance imaging technique, has enabled the characterization of the microstrucutral organization of tissue in vivo. As the brain develops, the water content in the brain decreases while protein and fat content increases due to processes such as myelination and axonal organization. Changes of signal intensity in structural MRI and diffusion parameters of DTI reflect these underlying biological changes.
Longitudinal neuroimaging studies provide a unique opportunity for understanding brain maturation by taking repeated scans over a time course within individuals. Despite the availability of detailed images of the brain, there has been little progress in accurate modeling of brain development or creating predictive models of structure that could help identify early signs of illness. We have developed methodologies for the nonlinear parametric modeling of longitudinal structural MRI and DTI changes over the neurodevelopmental period to address this gap. This research provides a normative model of early brain growth trajectory as is represented in structural MRI and DTI data, which will be crucial to understanding the timing and potential mechanisms of atypical development. Growth trajectories are described via intuitive parameters related to delay, rate of growth and expected asymptotic values, all descriptive measures that can answer clinical questions related to quantitative analysis of growth patterns. We demonstrate the potential of the framework on two clinical studies: healthy controls (singletons and twins) and children at risk of autism. Our framework is designed not only to provide qualitative comparisons, but also to give researchers and clinicians quantitative parameters and a statistical testing scheme. Moreover, the method includes modeling of growth trajectories of individuals, resulting in personalized profiles. The statistical framework also allows for prediction and prediction intervals for subject-specific growth trajectories, which will be crucial for efforts to improve diagnosis for individuals and personalized treatment.
Keywords: autism, brain development, image analysis


Nazmus Saquib.
“Visualizing Intrinsic Isosurface Variation due to Uncertainty Through Heat Kernel Signatures,” Note: Master of Science Thesis, Computational Engineering and Science (CES) Program, University of Utah School of Computing, 2013.
It is common to extract isosurfaces from simulation field data to visualize and gain understanding of the underlying physical phenomenon being simulated. As the input parameters of the simulation change, the resulting isosurface varies, and there has been increased interest in quantifying and visualization of these variations as part of the larger interest in uncertainty quantification. In this thesis, we propose an analysis and visualization pipeline for examining the intrinsic variation in isosurfaces caused by simulation parameter perturbation. Drawing inspiration from the shape modeling community, we incorporate the use of heat-kernel signatures (HKS) with a simple finite-difference approach for quantifying the degree to which a region (or even a point) on an isosurface has undergone intrinsic change. Coupled with a clustering technique and the use of color maps, our pipeline allows the user to select the level of fidelity with which they wish to evaluate and visualize the amount of intrinsic change. The pipeline is described with a simple example to walk the reader through the different steps, and experimental validation of parameter choices in the pipeline is provided to justify our design. Then we present canonical and simulation examples to demonstrate the pipeline's use in different applications.


M. Schott, T. Martin, A.V.P. Grosset, S.T. Smith, C.D. Hansen.
“Ambient Occlusion Effects for Combined Volumes and Tubular Geometry,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 19, No. 6, Note: Selected as Spotlight paper for June 2013 issue, pp. 913--926. 2013.
DOI: 10.1109/TVCG.2012.306
This paper details a method for interactive direct volume rendering that computes ambient occlusion effects for visualizations that combine both volumetric and geometric primitives, specifically tube shaped geometric objects representing streamlines, magnetic field lines or DTI fiber tracts. The algorithm extends the recently presented Directional Occlusion Shading model to allow the rendering of those geometric shapes in combination with a context providing 3D volume, considering mutual occlusion between structures represented by a volume or geometry. Stream tube geometries are computed using an effective spline based interpolation and approximation scheme that avoids self intersection and maintains coherent orientation of the stream tube segments to avoid surface deforming twists. Furthermore, strategies to reduce the geometric and specular aliasing of the stream tubes are discussed.
Keywords: Volume rendering, ambient occlusion, stream tubes


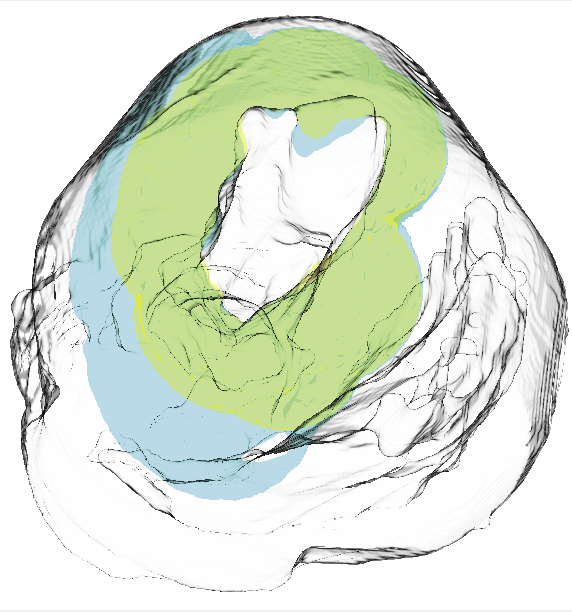
P.T. Scheffel, H.B. Henninger, R.T. Burks.
“Relationship of the intercondylar roof and the tibial footprint of the ACL: implications for ACL reconstruction,” In American Journal of Sports Medicine, Vol. 41, No. 2, pp. 396--401. 2013.
DOI: 10.1177/0363546512467955
Background: Debate exists on the proper relation of the anterior cruciate ligament (ACL) footprint with the intercondylar notch in anatomic ACL reconstructions. Patient-specific graft placement based on the inclination of the intercondylar roof has been proposed. The relationship between the intercondylar roof and native ACL footprint on the tibia has not previously been quantified.
Hypothesis: No statistical relationship exists between the intercondylar roof angle and the location of the native footprint of the ACL on the tibia.
Study Design: Case series; Level of evidence, 4.
Methods: Knees from 138 patients with both lateral radiographs and MRI, without a history of ligamentous injury or fracture, were reviewed to measure the intercondylar roof angle of the femur. Roof angles were measured on lateral radiographs. The MRI data of the same knees were analyzed to measure the position of the central tibial footprint of the ACL (cACL). The roof angle and tibial footprint were evaluated to determine if statistical relationships existed.
Results: Patients had a mean ± SD age of 40 ± 16 years. Average roof angle was 34.7° ± 5.2° (range, 23°-48°; 95% CI, 33.9°-35.5°), and it differed by sex but not by side (right/left). The cACL was 44.1% ± 3.4% (range, 36.1%-51.9%; 95% CI, 43.2%-45.0%) of the anteroposterior length of the tibia. There was only a weak correlation between the intercondylar roof angle and the cACL (R = 0.106). No significant differences arose between subpopulations of sex or side.
Conclusion: The tibial footprint of the ACL is located in a position on the tibia that is consistent and does not vary according to intercondylar roof angle. The cACL is consistently located between 43.2% and 45.0% of the anteroposterior length of the tibia. Intercondylar roof–based guidance may not predictably place a tibial tunnel in the native ACL footprint. Use of a generic ACL footprint to place a tibial tunnel during ACL reconstruction may be reliable in up to 95% of patients.


J. Schmidt, M. Berzins, J. Thornock, T. Saad, J. Sutherland.
“Large Scale Parallel Solution of Incompressible Flow Problems using Uintah and hypre,” In 2013 13th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGrid), pp. 458--465. 2013.
The Uintah Software framework was developed to provide an environment for solving fluid-structure interaction problems on structured adaptive grids on large-scale, longrunning, data-intensive problems. Uintah uses a combination of fluid-flow solvers and particle-based methods for solids together with a novel asynchronous task-based approach with fully automated load balancing. As Uintah is often used to solve incompressible flow problems in combustion applications it is important to have a scalable linear solver. While there are many such solvers available, the scalability of those codes varies greatly. The hypre software offers a range of solvers and preconditioners for different types of grids. The weak scalability of Uintah and hypre is addressed for particular examples of both packages when applied to a number of incompressible flow problems. After careful software engineering to reduce startup costs, much better than expected weak scalability is seen for up to 100K cores on NSFs Kraken architecture and up to 260K cpu cores, on DOEs new Titan machine. The scalability is found to depend in a crtitical way on the choice of algorithm used by hypre for a realistic application problem.
Keywords: Uintah, hypre, parallelism, scalability, linear equations


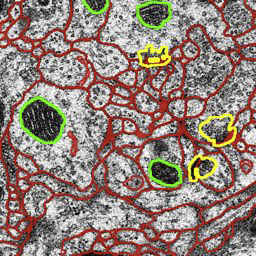
M. Seyedhosseini, M. Ellisman, T. Tasdizen.
“Segmentation of Mitochondria in Electron Microscopy Images using Algebraic Curves,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 860--863. 2013.
DOI: 10.1109/ISBI.2013.6556611
High-resolution microscopy techniques have been used to generate large volumes of data with enough details for understanding the complex structure of the nervous system. However, automatic techniques are required to segment cells and intracellular structures in these multi-terabyte datasets and make anatomical analysis possible on a large scale. We propose a fully automated method that exploits both shape information and regional statistics to segment irregularly shaped intracellular structures such as mitochondria in electron microscopy (EM) images. The main idea is to use algebraic curves to extract shape features together with texture features from image patches. Then, these powerful features are used to learn a random forest classifier, which can predict mitochondria locations precisely. Finally, the algebraic curves together with regional information are used to segment the mitochondria at the predicted locations. We demonstrate that our method outperforms the state-of-the-art algorithms in segmentation of mitochondria in EM images.


M. Seyedhosseini, T. Tasdizen.
“Multi-class Multi-scale Series Contextual Model for Image Segmentation,” In IEEE Transactions on Image Processing, Vol. PP, No. 99, 2013.
DOI: 10.1109/TIP.2013.2274388
Contextual information has been widely used as a rich source of information to segment multiple objects in an image. A contextual model utilizes the relationships between the objects in a scene to facilitate object detection and segmentation. However, using contextual information from different objects in an effective way for object segmentation remains a difficult problem. In this paper, we introduce a novel framework, called multi-class multi-scale (MCMS) series contextual model, which uses contextual information from multiple objects and at different scales for learning discriminative models in a supervised setting. The MCMS model incorporates cross-object and inter-object information into one probabilistic framework and thus is able to capture geometrical relationships and dependencies among multiple objects in addition to local information from each single object present in an image. We demonstrate that our MCMS model improves object segmentation performance in electron microscopy images and provides a coherent segmentation of multiple objects. By speeding up the segmentation process, the proposed method will allow neurobiologists to move beyond individual specimens and analyze populations paving the way for understanding neurodegenerative diseases at the microscopic level.


M. Seyedhosseini, M. Sajjadi, T. Tasdizen.
“Image Segmentation with Cascaded Hierarchical Models and Logistic Disjunctive Normal Networks,” In Proceedings of the IEEE International Conference on Computer Vison (ICCV 2013), pp. (accepted). 2013.
Contextual information plays an important role in solving vision problems such as image segmentation. However, extracting contextual information and using it in an effective way remains a difficult problem. To address this challenge, we propose a multi-resolution contextual framework, called cascaded hierarchical model (CHM), which learns contextual information in a hierarchical framework for image segmentation. At each level of the hierarchy, a classifier is trained based on downsampled input images and outputs of previous levels. Our model then incorporates the resulting multi-resolution contextual information into a classifier to segment the input image at original resolution. We repeat this procedure by cascading the hierarchical framework to improve the segmentation accuracy. Multiple classifiers are learned in the CHM; therefore, a fast and accurate classifier is required to make the training tractable. The classifier also needs to be robust against overfitting due to the large number of parameters learned during training. We introduce a novel classification scheme, called logistic disjunctive normal networks (LDNN), which consists of one adaptive layer of feature detectors implemented by logistic sigmoid functions followed by two fixed layers of logical units that compute conjunctions and disjunctions, respectively. We demonstrate that LDNN outperforms state-of-theart classifiers and can be used in the CHM to improve object segmentation performance.


A. Sharma, P.T. Fletcher, J.H. Gilmore, M.L. Escolar, A. Gupta, M. Styner, G. Gerig.
“Spatiotemporal Modeling of Discrete-Time Distribution-Valued Data Applied to DTI Tract Evolution in Infant Neurodevelopment,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 684--687. 2013.
DOI: 10.1109/ISBI.2013.6556567
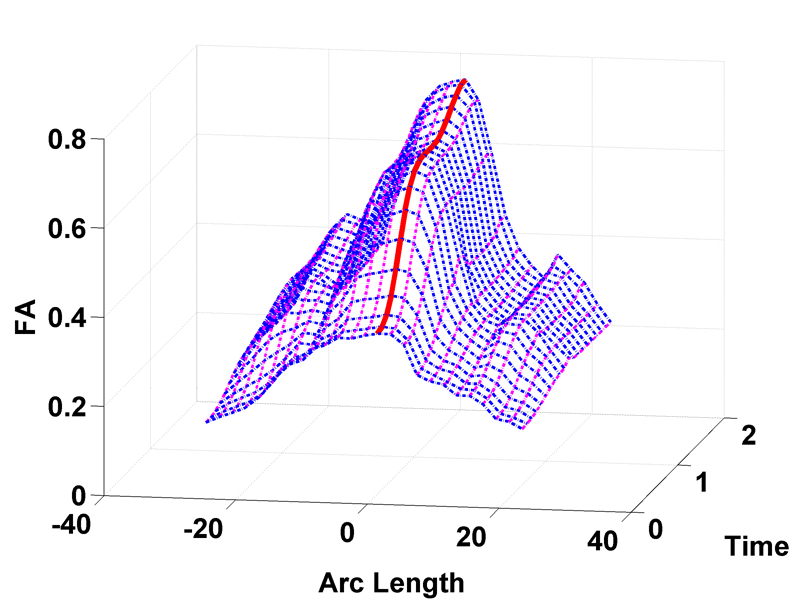
This paper proposes a novel method that extends spatiotemporal growth modeling to distribution-valued data. The method relaxes assumptions on the underlying noise models by considering the data to be represented by the complete probability distributions rather than a representative, single-valued summary statistics like the mean. When summarizing by the latter method, information on the underlying variability of data is lost early in the process and is not available at later stages of statistical analysis. The concept of ’distance’ between distributions and an ’average’ of distributions is employed. The framework quantifies growth trajectories for individuals and populations in terms of the complete data variability estimated along time and space. Concept is demonstrated in the context of our driving application which is modeling of age-related changes along white matter tracts in early neurodevelopment. Results are shown for a single subject with Krabbe's disease in comparison with a normative trend estimated from 15 healthy controls.


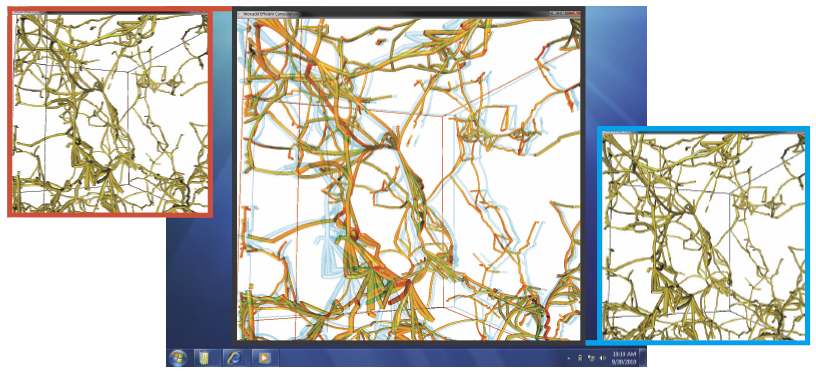
Y. Shi, G. Roger, C. Vachet, F. Budin, E. Maltbie, A. Verde, M. Hoogstoel, J.-B. Berger, M. Styner.
“Software-based diffusion MR human brain phantom for evaluating fiber-tracking algorithms,” In Proceedings of SPIE 8669, Medical Imaging 2013: Image Processing, 86692A, 2013.
DOI: 10.1117/12.2006113
PubMed ID: 24357914
PubMed Central ID: PMC3865235
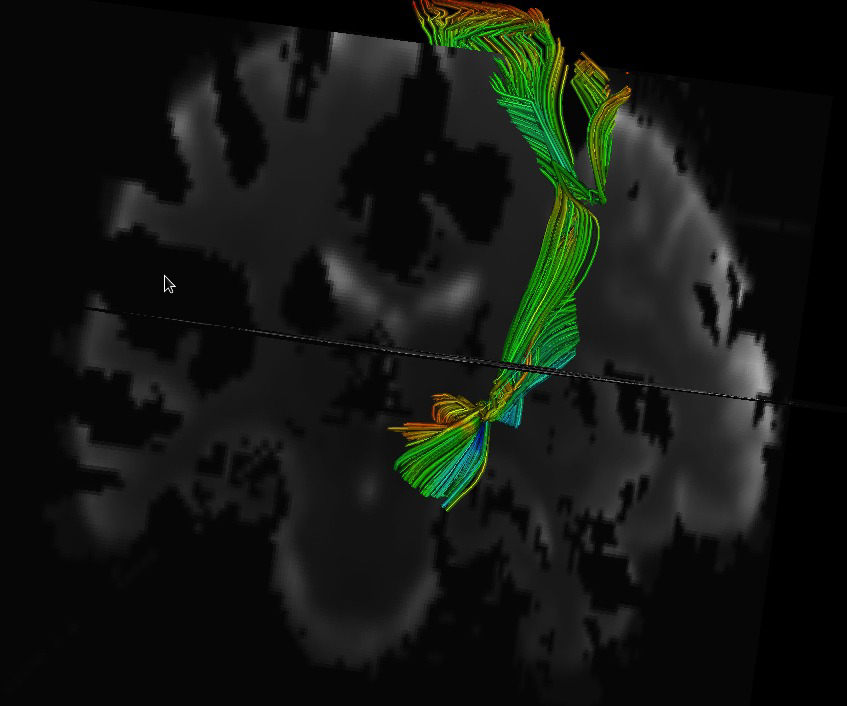
Fiber tracking provides insights into the brain white matter network and has become more and more popular in diffusion magnetic resonance (MR) imaging. Hardware or software phantom provides an essential platform to investigate, validate and compare various tractography algorithms towards a "gold standard". Software phantoms excel due to their flexibility in varying imaging parameters, such as tissue composition, SNR, as well as potential to model various anatomies and pathologies. This paper describes a novel method in generating diffusion MR images with various imaging parameters from realistically appearing, individually varying brain anatomy based on predefined fiber tracts within a high-resolution human brain atlas. Specifically, joint, high resolution DWI and structural MRI brain atlases were constructed with images acquired from 6 healthy subjects (age 22-26) for the DWI data and 56 healthy subject (age 18-59) for the structural MRI data. Full brain fiber tracking was performed with filtered, two-tensor tractography in atlas space. A deformation field based principal component model from the structural MRI as well as unbiased atlas building was then employed to generate synthetic structural brain MR images that are individually varying. Atlas fiber tracts were accordingly warped into each synthetic brain anatomy. Diffusion MR images were finally computed from these warped tracts via a composite hindered and restricted model of diffusion with various imaging parameters for gradient directions, image resolution and SNR. Furthermore, an open-source program was developed to evaluate the fiber tracking results both qualitatively and quantitatively based on various similarity measures.


S. Short, J.T. Elison, B.D. Goldman, M. Styner, H. Gu, M. Connelly, E. Maltbie, S. Woolson, W. Lin, G. Gerig, J.S. Reznick, J.H. Gilmore.
“Associations Between White Matter Microstructure and Infants' Working Memory,” In Neuroimage, Vol. 64, No. 1, Elsvier, pp. 156--166. January, 2013.
DOI: 10.1016/j.neuroimage.2012.09.021
PubMed ID: 22989623
Working memory emerges in infancy and plays a privileged role in subsequent adaptive cognitive development. The neural networks important for the development of working memory during infancy remain unknown. We used diffusion tensor imaging (DTI) and deterministic fiber tracking to characterize the microstructure of white matter fiber bundles hypothesized to support working memory in 12-month-old infants (n=73). Here we show robust associations between infants' visuospatial working memory performance and microstructural characteristics of widespread white matter. Significant associations were found for white matter tracts that connect brain regions known to support working memory in older children and adults (genu, anterior and superior thalamic radiations, anterior cingulum, arcuate fasciculus, and the temporal-parietal segment). Better working memory scores were associated with higher FA and lower RD values in these selected white matter tracts. These tract-specific brain-behavior relationships accounted for a significant amount of individual variation above and beyond infants' gestational age and developmental level, as measured with the Mullen Scales of Early Learning. Working memory was not associated with global measures of brain volume, as expected, and few associations were found between working memory and control white matter tracts. To our knowledge, this study is among the first demonstrations of brain-behavior associations in infants using quantitative tractography. The ability to characterize subtle individual differences in infant brain development associated with complex cognitive functions holds promise for improving our understanding of normative development, biomarkers of risk, experience-dependent learning and neuro-cognitive periods of developmental plasticity.

S.C. Sibole, S.A. Maas, J.P. Halloran, J.A. Weiss, A. Erdemir.
“Evaluation of a post-processing approach for multiscale analysis of biphasic mechanics of chondrocytes,” In Computer Methods in Biomechanical and Biomedical Engineering, Vol. 16, No. 10, pp. 1112--1126. 2013.
DOI: 10.1080/10255842.2013.809711
PubMed ID: 23809004
Understanding the mechanical behaviour of chondrocytes as a result of cartilage tissue mechanics has significant implications for both evaluation of mechanobiological function and to elaborate on damage mechanisms. A common procedure for prediction of chondrocyte mechanics (and of cell mechanics in general) relies on a computational post-processing approach where tissue-level deformations drive cell-level models. Potential loss of information in this numerical coupling approach may cause erroneous cellular-scale results, particularly during multiphysics analysis of cartilage. The goal of this study was to evaluate the capacity of first- and second-order data passing to predict chondrocyte mechanics by analysing cartilage deformations obtained for varying complexity of loading scenarios. A tissue-scale model with a sub-region incorporating representation of chondron size and distribution served as control. The post-processing approach first required solution of a homogeneous tissue-level model, results of which were used to drive a separate cell-level model (same characteristics as the sub-region of control model). The first-order data passing appeared to be adequate for simplified loading of the cartilage and for a subset of cell deformation metrics, for example, change in aspect ratio. The second-order data passing scheme was more accurate, particularly when asymmetric permeability of the tissue boundaries was considered. Yet, the method exhibited limitations for predictions of instantaneous metrics related to the fluid phase, for example, mass exchange rate. Nonetheless, employing higher order data exchange schemes may be necessary to understand the biphasic mechanics of cells under lifelike tissue loading states for the whole time history of the simulation.