SCI Publications
2013


N.P. Singh, J. Hinkle, S. Joshi, P.T. Fletcher.
“A Vector Momenta Formulation of Diffeomorphisms for Improved Geodesic Regression and Atlas Construction,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), Note: Received Best Student Paper Award, pp. 1219--1222. 2013.
DOI: 10.1109/ISBI.2013.6556700
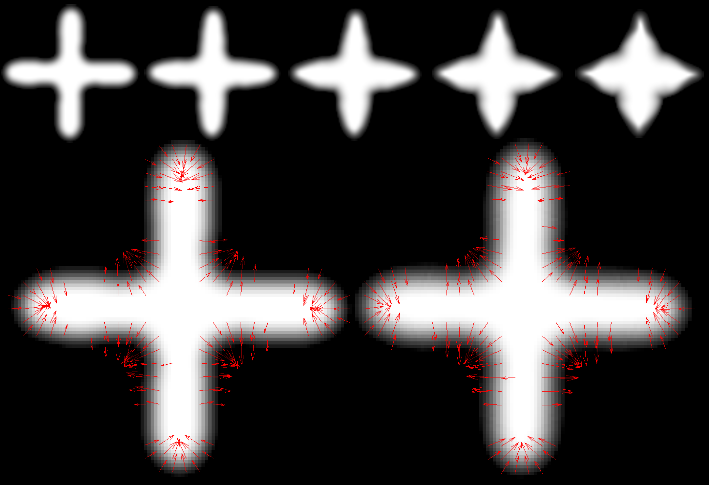
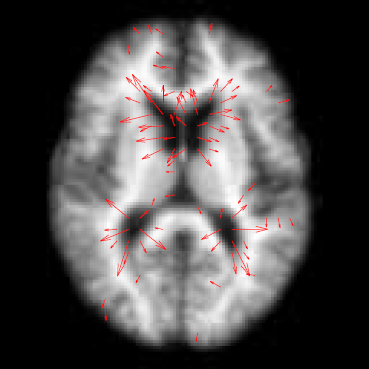
This paper presents a novel approach for diffeomorphic image regression and atlas estimation that results in improved convergence and numerical stability. We use a vector momenta representation of a diffeomorphism's initial conditions instead of the standard scalar momentum that is typically used. The corresponding variational problem results in a closed form update for template estimation in both the geodesic regression and atlas estimation problems. While we show that the theoretical optimal solution is equivalent to the scalar momenta case, the simplification of the optimization problem leads to more stable and efficient estimation in practice. We demonstrate the effectiveness of our method for atlas estimation and geodesic regression using synthetically generated shapes and 3D MRI brain scans.
Keywords: LDDMM, Geodesic regression, Atlas, Vector Momentum


N.P. Singh, J. Hinkle, S. Joshi, P.T. Fletcher.
“A Hierarchical Geodesic Model for Diffeomorphic Longitudinal Shape Analysis,” In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI), Lecture Notes in Computer Science (LNCS), pp. (accepted). 2013.
Hierarchical linear models (HLMs) are a standard approach for analyzing data where individuals are measured repeatedly over time. However, such models are only applicable to longitudinal studies of Euclidean data. In this paper, we propose a novel hierarchical geodesic model (HGM), which generalizes HLMs to the manifold setting. Our proposed model explains the longitudinal trends in shapes represented as elements of the group of diffeomorphisms. The individual level geodesics represent the trajectory of shape changes within individuals. The group level geodesic represents the average trajectory of shape changes for the population. We derive the solution of HGMs on diffeomorphisms to estimate individual level geodesics, the group geodesic, and the residual geodesics. We demonstrate the effectiveness of HGMs for longitudinal analysis of synthetically generated shapes and 3D MRI brain scans.


P. Skraba, Bei Wang, G. Chen, P. Rosen.
“2D Vector Field Simplification Based on Robustness,” SCI Technical Report, No. UUSCI-2013-004, SCI Institute, University of Utah, 2013.
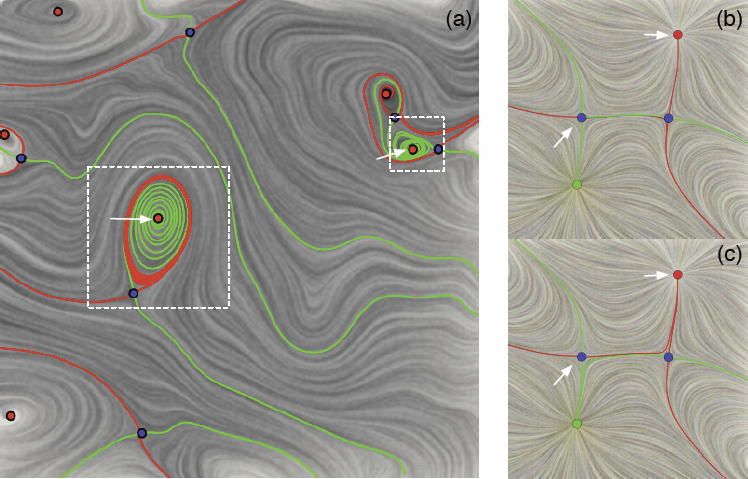
Vector field simplification aims to reduce the complexity of the flow by removing features in order of their relevance and importance, to reveal prominent behavior and obtain a compact representation for interpretation. Most existing simplification techniques based on the topological skeleton successively remove pairs of critical points connected by separatrices using distance or area-based relevance measures. These methods rely on the stable extraction of the topological skeleton, which can be difficult due to instability in numerical integration, especially when processing highly rotational flows. Further, the distance and area-based metrics are used to determine the cancellation ordering of features from a geometric point of view. Specifically, these metrics do not consider the flow magnitude, which is an important physical property of the flow. In this paper, we propose a novel simplification scheme derived from the recently introduced topological notion of robustness, which provides a complementary flow structure hierarchy to the traditional topological skeleton-based approach. Robustness enables the pruning of sets of critical points according to a quantitative measure of their stability, that is, the minimum amount of vector field perturbation required to remove them within a local neighborhood. This leads to a natural hierarchical simplification scheme with more physical consideration than purely topological-skeleton-based methods. Such a simplification does not depend on the topological skeleton of the vector field and therefore can handle more general situations (e.g. centers and pairs not connected by separatrices). We also provide a novel simplification algorithm based on degree theory with fewer restrictions and so can handle more general boundary conditions. We provide an implementation under the piecewise-linear setting and apply it to both synthetic and real-world datasets.


J. Sourati, D.H. Brooks, J.G. Dy, E. Erdogmus.
“Constrained Spectral Clustering for Image Segmentation,” In IEEE International Workshop on Machine Learning for Signal Processing, pp. 1--6. 2013.
DOI: 10.1109/MLSP
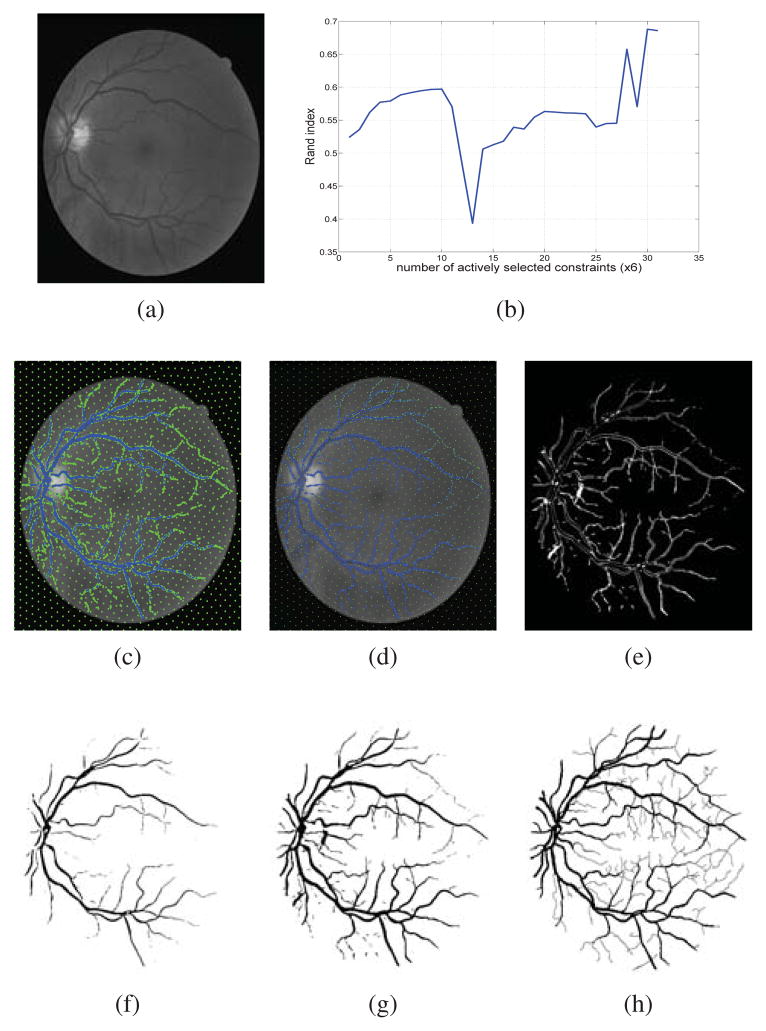
Constrained spectral clustering with affinity propagation in its original form is not practical for large scale problems like image segmentation. In this paper we employ novelty selection sub-sampling strategy, besides using efficient numerical eigen-decomposition methods to make this algorithm work efficiently for images. In addition, entropy-based active learning is also employed to select the queries posed to the user more wisely in an interactive image segmentation framework. We evaluate the algorithm on general and medical images to show that the segmentation results will improve using constrained clustering even if one works with a subset of pixels. Furthermore, this happens more efficiently when pixels to be labeled are selected actively.

J. Sourati, K. Kose, M. Rajadhyaksha, J.G. Dy, D. Erdogmus, D.H. Brooks.
“Automated localization of wrinkles and the dermo-epidermal junction in obliquely oriented reflectance confocal microscopic images of human skin,” In Proc. SPIE 8565, Photonic Therapeutics and Diagnostics IX, Vol. 8565, 2013.
DOI: 10.1117/12.2006489
Reflectance Confocal Microscopic (RCM) imaging of obliquely-oriented optical sections, rather than with traditional z-stacks, shows depth information that more closely mimics the appearance of skin in orthogonal sections of histology. This approach may considerably reduce the amount of data that must be acquired and processed. However, as with z-stacks, purely visual detection of the dermal-epidermal junction (DEJ) in oblique images remains challenging. Here, we have extended our original algorithm for localization of DEJ in z-stacks to oblique images. In addition, we developed an algorithm for detecting wrinkles, which in addition to its intrinsic merit, gives useful information for DEJ detection.


W.C. Stacey, S. Kellis, B. Greger, C.R. Butson, P.R. Patel, T. Assaf, T. Mihaylova, S. Glynn.
“Potential for unreliable interpretation of EEG recorded with microelectrodes,” In Epilepsia, May, 2013.
ISSN: 00139580
DOI: 10.1111/epi.12202


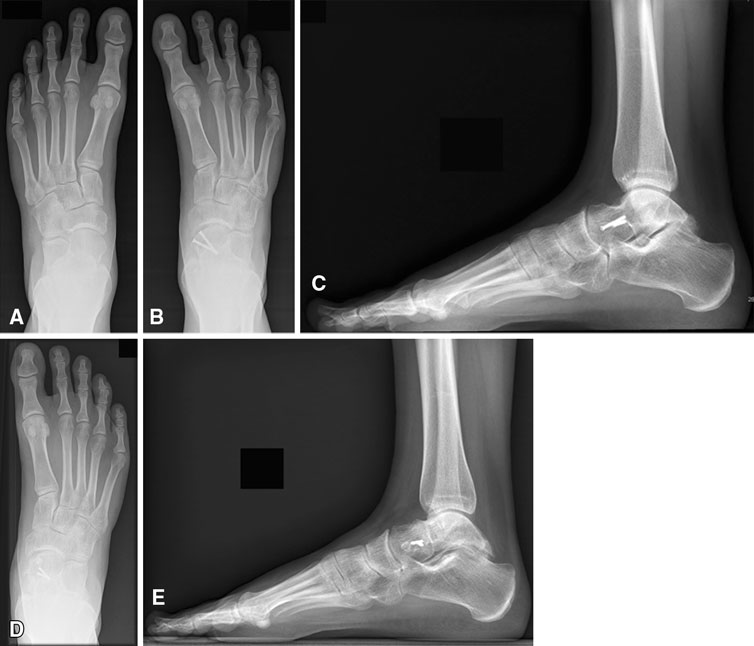
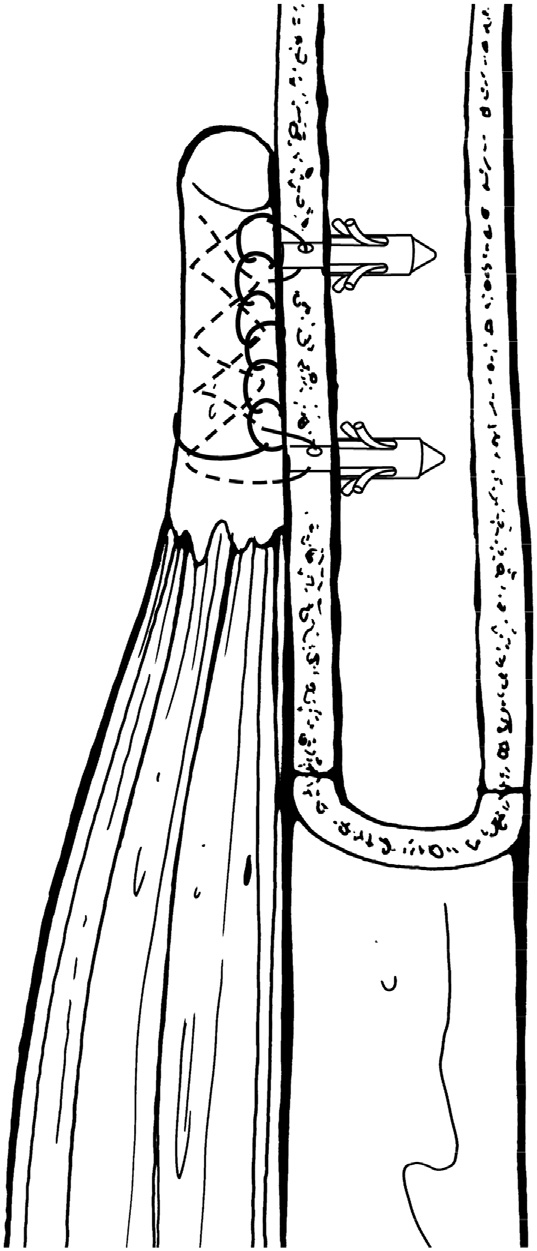
T. Suter, A. Barg, M. Knupp, H.B. Henninger, B. Hintermann.
“Surgical technique: talar neck osteotomy to lengthen the medial column after a malunited talar neck fracture,” In Clinical Orthopaedics & Related research, Vol. 471, No. 4, pp. 1356--1364. 2013.
DOI: 10.1080/10255842.2013.809711
PubMed ID: 23809004
Understanding the mechanical behaviour of chondrocytes as a result of cartilage tissue mechanics has significant implications for both evaluation of mechanobiological function and to elaborate on damage mechanisms. A common procedure for prediction of chondrocyte mechanics (and of cell mechanics in general) relies on a computational post-processing approach where tissue-level deformations drive cell-level models. Potential loss of information in this numerical coupling approach may cause erroneous cellular-scale results, particularly during multiphysics analysis of cartilage. The goal of this study was to evaluate the capacity of first- and second-order data passing to predict chondrocyte mechanics by analysing cartilage deformations obtained for varying complexity of loading scenarios. A tissue-scale model with a sub-region incorporating representation of chondron size and distribution served as control. The post-processing approach first required solution of a homogeneous tissue-level model, results of which were used to drive a separate cell-level model (same characteristics as the sub-region of control model). The first-order data passing appeared to be adequate for simplified loading of the cartilage and for a subset of cell deformation metrics, for example, change in aspect ratio. The second-order data passing scheme was more accurate, particularly when asymmetric permeability of the tissue boundaries was considered. Yet, the method exhibited limitations for predictions of instantaneous metrics related to the fluid phase, for example, mass exchange rate. Nonetheless, employing higher order data exchange schemes may be necessary to understand the biphasic mechanics of cells under lifelike tissue loading states for the whole time history of the simulation.


M. Szegedi, J. Hinkle, P. Rassiah, V. Sarkar, B. Wang, S. Joshi, B. Salter.
“Four‐dimensional tissue deformation reconstruction (4D TDR) validation using a real tissue phantom,” In Journal of Applied Clinical Medical Physics, Vol. 14, No. 1, pp. 115-132. 2013.
DOI: 10.1120/jacmp.v14i1.4012
Calculation of four‐dimensional (4D) dose distributions requires the remapping of dose calculated on each available binned phase of the 4D CT onto a reference phase for summation. Deformable image registration (DIR) is usually used for this task, but unfortunately almost always considers only endpoints rather than the whole motion path. A new algorithm, 4D tissue deformation reconstruction (4D TDR), that uses either CT projection data or all available 4D CT images to reconstruct 4D motion data, was developed. The purpose of this work is to verify the accuracy of the fit of this new algorithm using a realistic tissue phantom. A previously described fresh tissue phantom with implanted electromagnetic tracking (EMT) fiducials was used for this experiment. The phantom was animated using a sinusoidal and a real patient‐breathing signal. Four‐dimensional computer tomography (4D CT) and EMT tracking were performed. Deformation reconstruction was conducted using the 4D TDR and a modified 4D TDR which takes real tissue hysteresis (4D TDRHysteresis) into account. Deformation estimation results were compared to the EMT and 4D CT coordinate measurements. To eliminate the possibility of the high contrast markers driving the 4D TDR, a comparison was made using the original 4D CT data and data in which the fiducials were electronically masked. For the sinusoidal animation, the average deviation of the 4D TDR compared to the manually determined coordinates from 4D CT data was 1.9 mm, albeit with as large as 4.5 mm deviation. The 4D TDR calculation traces matched 95% of the EMT trace within 2.8 mm. The motion hysteresis generated by real tissue is not properly projected other than at endpoints of motion. Sinusoidal animation resulted in 95% of EMT measured locations to be within less than 1.2 mm of the measured 4D CT motion path, enabling accurate motion characterization of the tissue hysteresis. The 4D TDRHysteresis calculation traces accounted well for the hysteresis and matched 95% of the EMT trace within 1.6 mm. An irregular (in amplitude and frequency) recorded patient trace applied to the same tissue resulted in 95% of the EMT trace points within less than 4.5 mm when compared to both the 4D CT and 4D TDRHysteresis motion paths. The average deviation of 4D TDRHysteresis compared to 4D CT datasets was 0.9 mm under regular sinusoidal and 1.0 mm under irregular patient trace animation. The EMT trace data fit to the 4D TDRHysteresis was within 1.6 mm for sinusoidal and 4.5 mm for patient trace animation. While various algorithms have been validated for end‐to‐end accuracy, one can only be fully confident in the performance of a predictive algorithm if one looks at data along the full motion path. The 4D TDR, calculating the whole motion path rather than only phase‐ or endpoints, allows us to fully characterize the accuracy of a predictive algorithm, minimizing assumptions. This algorithm went one step further by allowing for the inclusion of tissue hysteresis effects, a real‐world effect that is neglected when endpoint‐only validation is performed. Our results show that the 4D TDRHysteresis correctly models the deformation at the endpoints and any intermediate points along the motion path.
PACS numbers: 87.55.km, 87.55.Qr, 87.57.nf, 87.85.Tu


R.Z. Tashjian, H.B. Henninger.
“Biomechanical evaluation of subpectoral biceps tenodesis: dual suture anchor versus interference screw fixation,” In Journal of Shoulder and Elbow Surgery, Vol. 22, No. 10, pp. 1408–-1412. 2013.
DOI: 10.1016/j.jse.2012.12.039
Background
Subpectoral biceps tenodesis has been reliably used to treat a variety of biceps tendon pathologies. Interference screws have been shown to have superior biomechanical properties compared to suture anchors; although, only single anchor constructs have been evaluated in the subpectoral region. The purpose of this study was to compare interference screw fixation with a suture anchor construct, using 2 anchors for a subpectoral tenodesis.
Methods
A subpectoral biceps tenodesis was performed using either an interference screw (8 × 12 mm; Arthrex) or 2 suture anchors (Mitek G4) with #2 FiberWire (Arthrex) in a Krackow and Bunnell configuration in seven pairs of human cadavers. The humerus was inverted in an Instron and the biceps tendon was loaded vertically. Displacement driven cyclic loading was performed followed by failure loading.
Results
Suture anchor constructs had lower stiffness upon initial loading (P = .013). After 100 cycles, the stiffness of the suture anchor construct "softened" (decreased 9%, P < .001), whereas the screw construct was unchanged (0.4%, P = .078). Suture anchors had significantly higher ultimate failure strain than the screws (P = .003), but ultimate failure loads were similar between constructs: 280 ± 95 N (screw) vs 310 ± 91 N (anchors) (P = .438).
Conclusion
The interference screw was significantly stiffer than the suture anchor construct. Ultimate failure loads were similar between constructs, unlike previous reports indicating interference screws had higher ultimate failure loads compared to suture anchors. Neither construct was superior with regards to stress; although, suture anchors could withstand greater elongation prior to failure.


A. Vardhan, M.W. Prastawa, J. Piven, G. Gerig.
“Modeling Longitudinal MRI Changes in Populations Using a Localized, Information-Theoretic Measure of Contrast,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 1396--1399. 2013.
DOI: 10.1109/ISBI.2013.6556794
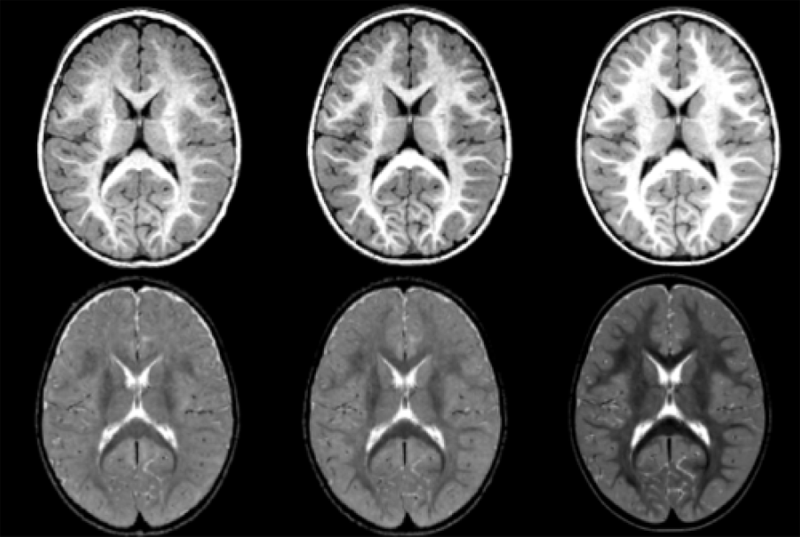
Longitudinal MR imaging during early brain development provides important information about growth patterns and the development of neurological disorders. We propose a new framework for studying brain growth patterns within and across populations based on MRI contrast changes, measured at each time point of interest and at each voxel. Our method uses regression in the LogOdds space and an informationtheoretic measure of distance between distributions to capture contrast in a manner that is robust to imaging parameters and without requiring intensity normalization. We apply our method to a clinical neuroimaging study on early brain development in autism, where we obtain a 4D spatiotemporal model of contrast changes in multimodal structural MRI.


A. Vardhan, J. Piven, M. Prastawa, G. Gerig.
“A longitudinal structural MRI study of change in regional contrast in Autism Spectrum Disorder,” In Proceedings of the 19th Annual Meeting of the Organization for Human Brain Mapping OHBM, pp. (in print). 2013.
The brain undergoes tremendous changes in shape, size, structure, and chemical composition, between birth and 2 years of age [Rutherford, 2001]. Existing studies have focused on morphometric and volumetric changes to study the early developing brain. Although there have been some recent appearance studies based on intensity changes [Serag et al., 2011], these are highly dependent on the quality of normalization. The study we present here uses the changes in contrast between gray and white matter tissue intensities in structural MRI of the brain, as a measure of regional growth [Vardhan et al., 2011]. Kernel regression was used to generate continuous curves characterizing the changes in contrast with time. A statistical analysis was then performed on these curves, comparing two population groups : (i) HR+ : high-risk subjects who tested positive for Autism Spectrum Disorder (ASD), and (ii) HR- : high-risk subjects who tested negative for ASD.


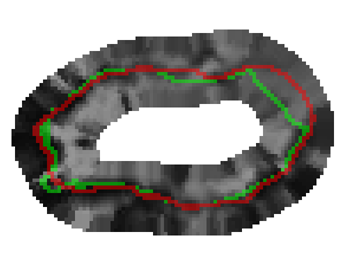
G. Veni, Z. Fu, S.P. Awate, R.T. Whitaker.
“Proper Ordered Meshing of Complex Shapes and Optimal Graph Cuts Applied to Atrial-Wall Segmentation from DE-MRI,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 1296--1299. 2013.
DOI: 10.1109/ISBI.2013.6556769
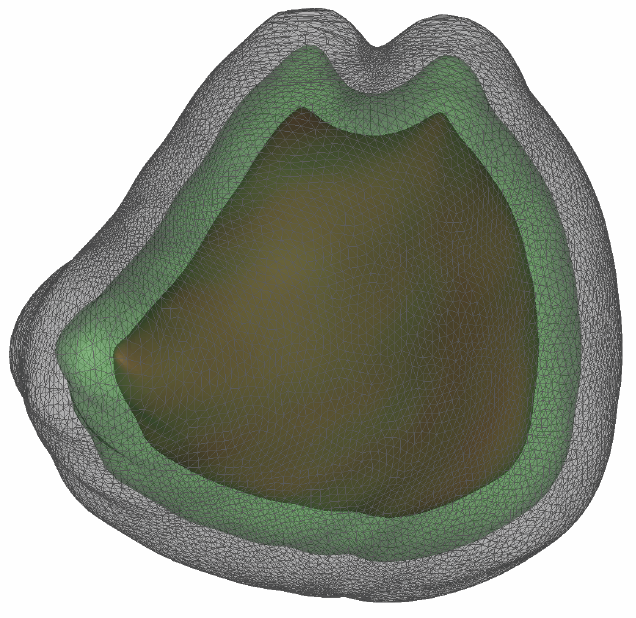
Segmentation of the left atrium wall from delayed enhancement MRI is challenging because of inconsistent contrast combined with noise and high variation in atrial shape and size. This paper presents a method for left-atrium wall segmentation by using a novel sophisticated mesh-generation strategy and graph cuts on a proper ordered graph. The mesh is part of a template/model that has an associated set of learned intensity features. When this mesh is overlaid onto a test image, it produces a set of costs on the graph vertices which eventually leads to an optimal segmentation. The novelty also lies in the construction of proper ordered graphs on complex shapes and for choosing among distinct classes of base shapes/meshes for automatic segmentation. We evaluate the proposed segmentation framework quantitatively on simulated and clinical cardiac MRI.


G. Veni, S. Awate, Z. Fu, R.T. Whittaker.
“Bayesian Segmentation of Atrium Wall using Globally-Optimal Graph Cuts on 3D Meshes,” In Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI), Lecture Notes in Computer Science (LNCS), Vol. 23, pp. 656--677. 2013.
PubMed ID: 24684007
Efficient segmentation of the left atrium (LA) wall from delayed enhancement MRI is challenging due to inconsistent contrast, combined with noise, and high variation in atrial shape and size. We present a surface-detection method that is capable of extracting the atrial wall by computing an optimal a-posteriori estimate. This estimation is done on a set of nested meshes, constructed from an ensemble of segmented training images, and graph cuts on an associated multi-column, proper-ordered graph. The graph/mesh is a part of a template/model that has an associated set of learned intensity features. When this mesh is overlaid onto a test image, it produces a set of costs which lead to an optimal segmentation. The 3D mesh has an associated weighted, directed multi-column graph with edges that encode smoothness and inter-surface penalties. Unlike previous graph-cut methods that impose hard constraints on the surface properties, the proposed method follows from a Bayesian formulation resulting in soft penalties on spatial variation of the cuts through the mesh. The novelty of this method also lies in the construction of proper-ordered graphs on complex shapes for choosing among distinct classes of base shapes for automatic LA segmentation. We evaluate the proposed segmentation framework on simulated and clinical cardiac MRI.
Keywords: Atrial Fibrillation, Bayesian segmentation, Minimum s-t cut, Mesh Generation, Geometric Graph


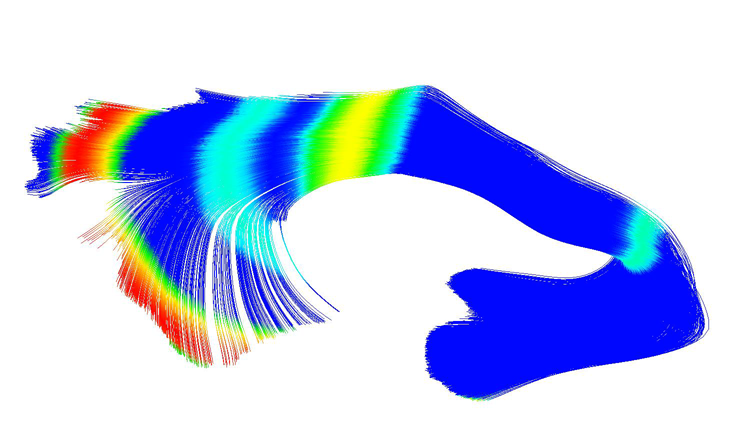
A.R. Verde, J.-B. Berger, A. Gupta, M. Farzinfar, A. Kaiser, V.W. Chanon, C. Boettiger, H. Johnson, J. Matsui, A. Sharma, C. Goodlett, Y. Shi, H. Zhu, G. Gerig, S. Gouttard, C. Vachet, M. Styner.
“UNC-Utah NA-MIC DTI framework: atlas based fiber tract analysis with application to a study of nicotine smoking addiction,” In Proc. SPIE 8669, Medical Imaging 2013: Image Processing, 86692D, Vol. 8669, pp. 86692D-86692D-8. 2013.
DOI: 10.1117/12.2007093
Purpose: The UNC-Utah NA-MIC DTI framework represents a coherent, open source, atlas fiber tract based DTI analysis framework that addresses the lack of a standardized fiber tract based DTI analysis workflow in the field. Most steps utilize graphical user interfaces (GUI) to simplify interaction and provide an extensive DTI analysis framework for non-technical researchers/investigators. Data: We illustrate the use of our framework on a 54 directional DWI neuroimaging study contrasting 15 Smokers and 14 Controls. Method(s): At the heart of the framework is a set of tools anchored around the multi-purpose image analysis platform 3D-Slicer. Several workflow steps are handled via external modules called from Slicer in order to provide an integrated approach. Our workflow starts with conversion from DICOM, followed by thorough automatic and interactive quality control (QC), which is a must for a good DTI study. Our framework is centered around a DTI atlas that is either provided as a template or computed directly as an unbiased average atlas from the study data via deformable atlas building. Fiber tracts are defined via interactive tractography and clustering on that atlas. DTI fiber profiles are extracted automatically using the atlas mapping information. These tract parameter profiles are then analyzed using our statistics toolbox (FADTTS). The statistical results are then mapped back on to the fiber bundles and visualized with 3D Slicer. Results: This framework provides a coherent set of tools for DTI quality control and analysis. Conclusions: This framework will provide the field with a uniform process for DTI quality control and analysis.


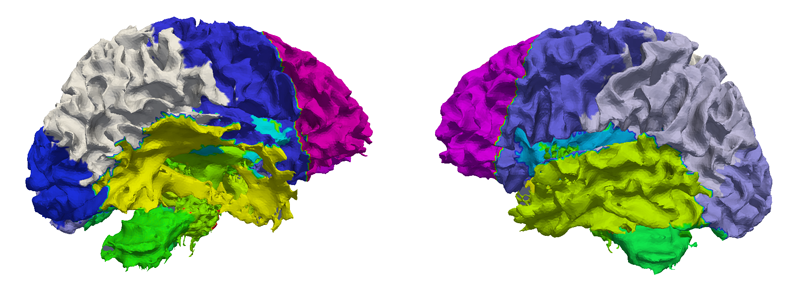
Bo Wang, M.W. Prastawa, A. Irimia, M.C. Chambers, N. Sadeghi, P.M. Vespa, J.D. van Horn, G. Gerig.
“Analyzing Imaging Biomarkers for Traumatic Brain Injury Using 4D Modeling of Longitudinal MRI,” In 2013 IEEE Proceedings of 10th International Symposium on Biomedical Imaging (ISBI), pp. 1392 - 1395. 2013.
DOI: 10.1109/ISBI.2013.6556793
Quantitative imaging biomarkers are important for assessment of impact, recovery and treatment efficacy in patients with traumatic brain injury (TBI). To our knowledge, the identification of such biomarkers characterizing disease progress and recovery has been insufficiently explored in TBI due to difficulties in registration of baseline and followup data and automatic segmentation of tissue and lesions from multimodal, longitudinal MR image data. We propose a new methodology for computing imaging biomarkers in TBI by extending a recently proposed spatiotemporal 4D modeling approach in order to compute quantitative features of tissue change. The proposed method computes surface-based and voxel-based measurements such as cortical thickness, volume changes, and geometric deformation. We analyze the potential for clinical use of these biomarkers by correlating them with TBI-specific patient scores at the level of the whole brain and of individual regions. Our preliminary results indicate that the proposed voxel-based biomarkers are correlated with clinical outcomes.


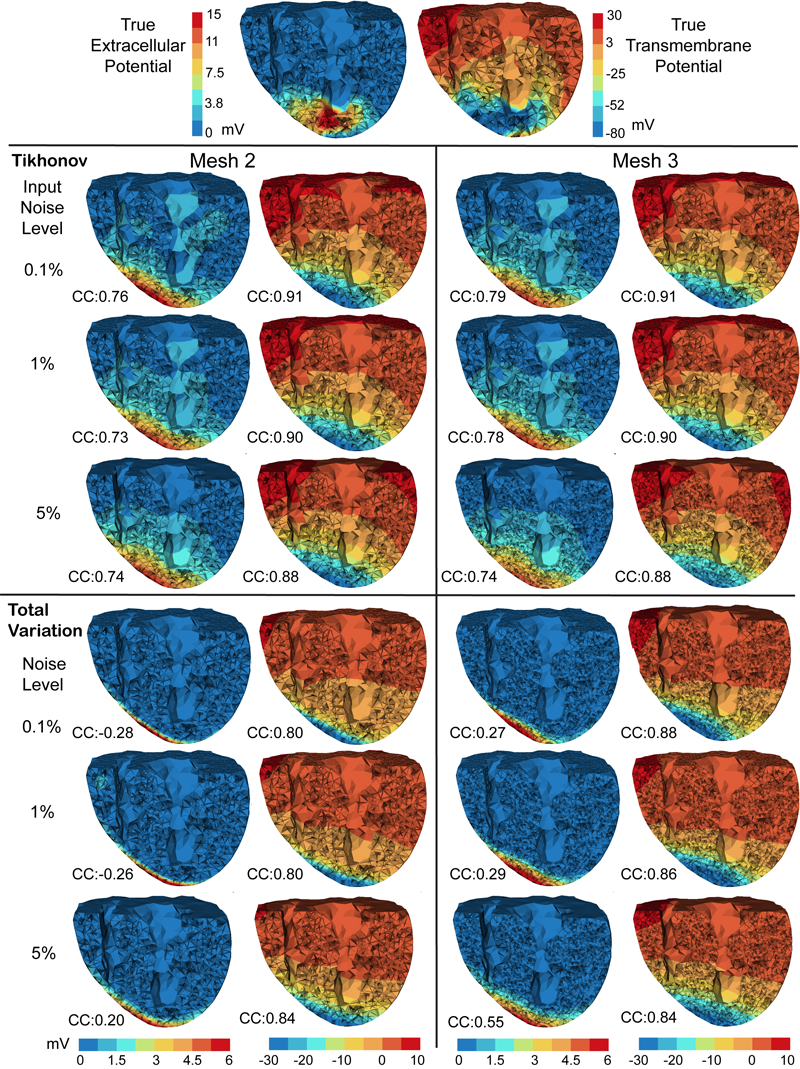
D. Wang, R.M. Kirby, R.S. MacLeod, C.R. Johnson.
“Inverse Electrocardiographic Source Localization of Ischemia: An Optimization Framework and Finite Element Solution,” In Journal of Computational Physics, Vol. 250, Academic Press, pp. 403--424. 2013.
ISSN: 0021-9991
DOI: 10.1016/j.jcp.2013.05.027
With the goal of non-invasively localizing cardiac ischemic disease using bodysurface potential recordings, we attempted to reconstruct the transmembrane potential (TMP) throughout the myocardium with the bidomain heart model. The task is an inverse source problem governed by partial differential equations (PDE). Our main contribution is solving the inverse problem within a PDE-constrained optimization framework that enables various physically-based constraints in both equality and inequality forms. We formulated the optimality conditions rigorously in the continuum before deriving finite element discretization, thereby making the optimization independent of discretization choice. Such a formulation was derived for the L2-norm Tikhonov regularization and the total variation minimization. The subsequent numerical optimization was fulfilled by a primal-dual interior-point method tailored to our problem's specific structure. Our simulations used realistic, fiberincluded heart models consisting of up to 18,000 nodes, much finer than any inverse models previously reported. With synthetic ischemia data we localized ischemic regions with roughly a 10% false-negative rate or a 20% false-positive rate under conditions up to 5% input noise. With ischemia data measured from animal experiments, we reconstructed TMPs with roughly 0.9 correlation with the ground truth. While precisely estimating the TMP in general cases remains an open problem, our study shows the feasibility of reconstructing TMP during the ST interval as a means of ischemia localization.
Keywords: cvrti, 2P41 GM103545-14


Bei Wang, P. Rosen, P. Skraba, H. Bhatia, V. Pascucci.
“Visualizing Robustness of Critical Points for 2D Time-Varying Vector Fields,” In Computer Graphics Forum, Vol. 32, No. 3, Wiley-Blackwell, pp. 221--230. jun, 2013.
DOI: 10.1111/cgf.12109
Analyzing critical points and their temporal evolutions plays a crucial role in understanding the behavior of vector fields. A key challenge is to quantify the stability of critical points: more stable points may represent more important phenomena or vice versa. The topological notion of robustness is a tool which allows us to quantify rigorously the stability of each critical point. Intuitively, the robustness of a critical point is the minimum amount of perturbation necessary to cancel it within a local neighborhood, measured under an appropriate metric. In this paper, we introduce a new analysis and visualization framework which enables interactive exploration of robustness of critical points for both stationary and time-varying 2D vector fields. This framework allows the end-users, for the first time, to investigate how the stability of a critical point evolves over time. We show that this depends heavily on the global properties of the vector field and that structural changes can correspond to interesting behavior. We demonstrate the practicality of our theories and techniques on several datasets involving combustion and oceanic eddy simulations and obtain some key insights regarding their stable and unstable features.


Y. Wan, H. Otsuna, C.D. Hansen.
“Synthetic Brainbows,” In Computer Graphics Forum, Vol. 32, No. 3pt4, Wiley-Blackwell, pp. 471--480. jun, 2013.
DOI: 10.1111/cgf.12134
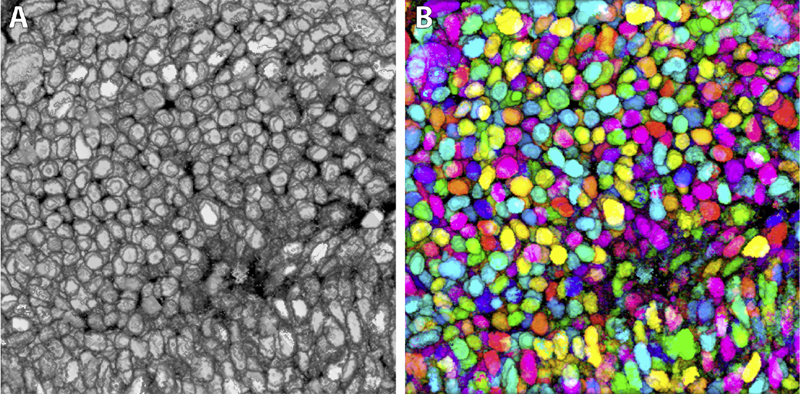
Brainbow is a genetic engineering technique that randomly colorizes cells. Biological samples processed with this technique and imaged with confocal microscopy have distinctive colors for individual cells. Complex cellular structures can then be easily visualized. However, the complexity of the Brainbow technique limits its applications. In practice, most confocal microscopy scans use different florescence staining with typically at most three distinct cellular structures. These structures are often packed and obscure each other in rendered images making analysis difficult. In this paper, we leverage a process known as GPU framebuffer feedback loops to synthesize Brainbow-like images. In addition, we incorporate ID shuffling and Monte-Carlo sampling into our technique, so that it can be applied to single-channel confocal microscopy data. The synthesized Brainbow images are presented to domain experts with positive feedback. A user survey demonstrates that our synthetic Brainbow technique improves visualizations of volume data with complex structures for biologists.


Bo Wang, M. Prastawa, A. Saha, S.P. Awate, A. Irimia, M.C. Chambers, P.M. Vespa, J.D. Van Horn, V. Pascucci, G. Gerig.
“Modeling 4D changes in pathological anatomy using domain adaptation: analysis of TBI imaging using a tumor database,” In Proceedings of the 2013 MICCAI-MBIA Workshop, Lecture Notes in Computer Science (LNCS), Vol. 8159, Note: Awarded Best Paper!, pp. 31--39. 2013.
DOI: 10.1007/978-3-319-02126-3_4
Analysis of 4D medical images presenting pathology (i.e., lesions) is signi cantly challenging due to the presence of complex changes over time. Image analysis methods for 4D images with lesions need to account for changes in brain structures due to deformation, as well as the formation and deletion of new structures (e.g., edema, bleeding) due to the physiological processes associated with damage, intervention, and recovery. We propose a novel framework that models 4D changes in pathological anatomy across time, and provides explicit mapping from a healthy template to subjects with pathology. Moreover, our framework uses transfer learning to leverage rich information from a known source domain, where we have a collection of completely segmented images, to yield effective appearance models for the input target domain. The automatic 4D segmentation method uses a novel domain adaptation technique for generative kernel density models to transfer information between different domains, resulting in a fully automatic method that requires no user interaction. We demonstrate the effectiveness of our novel approach with the analysis of 4D images of traumatic brain injury (TBI), using a synthetic tumor database as the source domain.


Y. Wan.
“Fluorender, An Interactive Tool for Confocal Microscopy Data Visualization and Analysis,” Note: Ph.D. Thesis, School of Computing, University of Utah, June, 2013.
Confocal microscopy has become a popular imaging technique in biology research in recent years. It is often used to study three-dimensional (3D) structures of biological samples. Confocal data are commonly multi-channel, with each channel resulting from a different fluorescent staining. This technique also results finely detailed structures in 3D, such as neuron fibers. Despite the plethora of volume rendering techniques that have been available for many years, there is a demand from biologists for a flexible tool that allows interactive visualization and analysis of multi-channel confocal data. Together with biologists, we have designed and developed FluoRender. It incorporates volume rendering techniques such as a two-dimensional (2D) transfer function and multi-channel intermixing. Rendering results can be enhanced through tone-mappings and overlays. To facilitate analyses of confocal data, FluoRender provides interactive operations for extracting complex structures. Furthermore, we developed the Synthetic Brainbow technique, which takes advantage of the asynchronous behavior in Graphics Processing Unit (GPU) framebuffer loops and generates random colorizations for different structures in single-channel confocal data. The results from our Synthetic Brainbows, when applied to a sequence of developing cells, can then be used for tracking the movements of these cells. Finally, we present an application of FluoRender in the workflow of constructing anatomical atlases.
Keywords: confocal microscopy, visualization, software