SCI Publications
2012


H.C. Hazlett, H. Gu, R.C. McKinstry, D.W.W. Shaw, K.N. Botteron, S. Dager, M. Styner, C. Vachet, G. Gerig, S. Paterson, R.T. Schultz, A.M. Estes, A.C. Evans, J. Piven.
“Brain Volume Findings in Six Month Old Infants at High Familial Risk for Autism,” In American Journal of Psychiatry (AJP), pp. (in print). 2012.
Objective: Brain enlargement has been observed in individuals with autism as early as two years of age. Studies using head circumference suggest that brain enlargement is a postnatal event that occurs around the latter part of the first year. To date, no brain imaging studies have systematically examined the period prior to age two. In this study we examine MRI brain volume in six month olds at high familial risk for autism.
Method: The Infant Brain Imaging Study (IBIS) is a longitudinal imaging study of infants at high risk for autism. This cross-sectional analysis examines brain volumes at six months of age, in high risk infants (N=98) in comparison to infants without family members with autism (low risk) (N=36). MRI scans are also examined for radiologic abnormalities.
Results: No group differences were observed for intracranial cerebrum, cerebellum, lateral ventricle volumes, or head circumference.
Conclusions: We did not observe significant group differences for head circumference, brain volume, or abnormalities of radiologic findings in a sample of 6 month old infants at highrisk for autism. We are unable to conclude that these changes are not present in infants who later go on to receive a diagnosis of autism, but rather that they were not detected in a large group at high familial risk. Future longitudinal studies of the IBIS sample will examine whether brain volume may differ in those infants who go onto develop autism, estimating that approximately 20\% of this sample may be diagnosed with an autism spectrum disorder at age two.


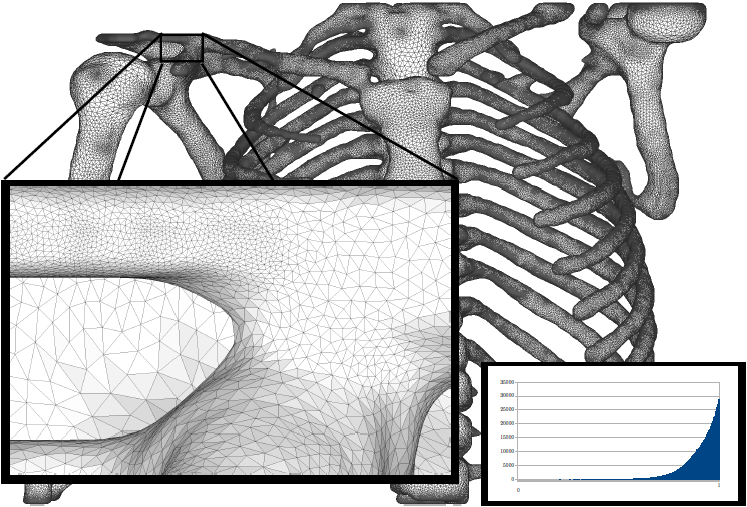
H.B. Henninger, Barg A, A.E. Anderson, K.N. Bachus, R.Z. Tashjian, R.T. Burks.
“Effect of deltoid tension and humeral version in reverse total shoulder arthroplasty: a biomechanical study,” In Journal of Shoulder and Elbow Surgery, Vol. 21, No. 4, pp. 483–-490. 2012.
DOI: 10.1016/j.jse.2011.01.040
Background
No clear recommendations exist regarding optimal humeral component version and deltoid tension in reverse total shoulder arthroplasty (TSA).
Materials and methods
A biomechanical shoulder simulator tested humeral versions (0°, 10°, 20° retroversion) and implant thicknesses (-3, 0, +3 mm from baseline) after reverse TSA in human cadavers. Abduction and external rotation ranges of motion as well as abduction and dislocation forces were quantified for native arms and arms implanted with 9 combinations of humeral version and implant thickness.
Results
Resting abduction angles increased significantly (up to 30°) after reverse TSA compared with native shoulders. With constant posterior cuff loads, native arms externally rotated 20°, whereas no external rotation occurred in implanted arms (20° net internal rotation). Humeral version did not affect rotational range of motion but did alter resting abduction. Abduction forces decreased 30% vs native shoulders but did not change when version or implant thickness was altered. Humeral center of rotation was shifted 17 mm medially and 12 mm inferiorly after implantation. The force required for lateral dislocation was 60% less than anterior and was not affected by implant thickness or version.
Conclusion
Reverse TSA reduced abduction forces compared with native shoulders and resulted in limited external rotation and abduction ranges of motion. Because abduction force was reduced for all implants, the choice of humeral version and implant thickness should focus on range of motion. Lateral dislocation forces were less than anterior forces; thus, levering and inferior/posterior impingement may be a more probable basis for dislocation (laterally) than anteriorly directed forces.
Keywords: Shoulder, reverse arthroplasty, deltoid tension, humeral version, biomechanical simulator


H.B. Henninger, A. Barg, A.E. Anderson, K.N. Bachus, R.T. Burks, R.Z. Tashjian.
“Effect of lateral offset center of rotation in reverse total shoulder arthroplasty: a biomechanical study,” In Journal of Shoulder and Elbow Surgery, Vol. 21, No. 9, pp. 1128--1135. 2012.
DOI: 10.1016/j.jse.2011.07.034
Background
Lateral offset center of rotation (COR) reduces the incidence of scapular notching and potentially increases external rotation range of motion (ROM) after reverse total shoulder arthroplasty (rTSA). The purpose of this study was to determine the biomechanical effects of changing COR on abduction and external rotation ROM, deltoid abduction force, and joint stability.
Materials and methods
A biomechanical shoulder simulator tested cadaveric shoulders before and after rTSA. Spacers shifted the COR laterally from baseline rTSA by 5, 10, and 15 mm. Outcome measures of resting abduction and external rotation ROM, and abduction and dislocation (lateral and anterior) forces were recorded.
Results
Resting abduction increased 20° vs native shoulders and was unaffected by COR lateralization. External rotation decreased after rTSA and was unaffected by COR lateralization. The deltoid force required for abduction significantly decreased 25% from native to baseline rTSA. COR lateralization progressively eliminated this mechanical advantage. Lateral dislocation required significantly less force than anterior dislocation after rTSA, and both dislocation forces increased with lateralization of the COR.
Conclusion
COR lateralization had no influence on ROM (adduction or external rotation) but significantly increased abduction and dislocation forces. This suggests the lower incidence of scapular notching may not be related to the amount of adduction deficit after lateral offset rTSA but may arise from limited impingement of the humeral component on the lateral scapula due to a change in joint geometry. Lateralization provides the benefit of increased joint stability, but at the cost of increasing deltoid abduction forces.
Keywords: Shoulder simulator, reverse arthroplasty, lateral offset, center of rotation


J. Hinkle, P. Muralidharan, P.T. Fletcher, S. Joshi.
“Polynomial Regression on Riemannian Manifolds,” In arXiv, Vol. 1201.2395, 2012.
In this paper we develop the theory of parametric polynomial regression in Riemannian manifolds and Lie groups. We show application of Riemannian polynomial regression to shape analysis in Kendall shape space. Results are presented, showing the power of polynomial regression on the classic rat skull growth data of Bookstein as well as the analysis of the shape changes associated with aging of the corpus callosum from the OASIS Alzheimer's study.


L. Hogrebe, A.R.C. Paiva, E. Jurrus, C. Christensen, M. Bridge, L. Dai, R.L. Pfeiffer, P.R. Hof, B. Roysam, J.R. Korenberg, T. Tasdizen.
“Serial section registration of axonal confocal microscopy datasets for long-range neural circuit reconstruction,” In Journal of Neuroscience Methods, Vol. 207, No. 2, pp. 200--210. 2012.
DOI: 10.1016/j.jneumeth.2012.03.002
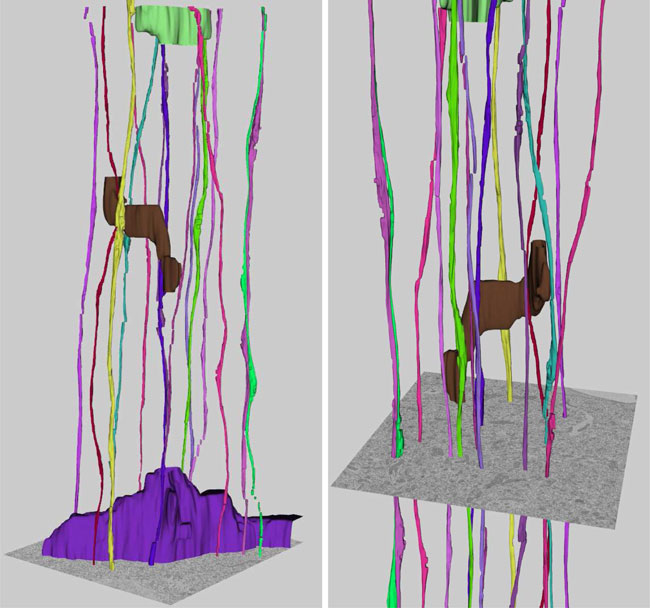
In the context of long-range digital neural circuit reconstruction, this paper investigates an approach for registering axons across histological serial sections. Tracing distinctly labeled axons over large distances allows neuroscientists to study very explicit relationships between the brain's complex interconnects and, for example, diseases or aberrant development. Large scale histological analysis requires, however, that the tissue be cut into sections. In immunohistochemical studies thin sections are easily distorted due to the cutting, preparation, and slide mounting processes. In this work we target the registration of thin serial sections containing axons. Sections are first traced to extract axon centerlines, and these traces are used to define registration landmarks where they intersect section boundaries. The trace data also provides distinguishing information regarding an axon's size and orientation within a section. We propose the use of these features when pairing axons across sections in addition to utilizing the spatial relationships among the landmarks. The global rotation and translation of an unregistered section are accounted for using a random sample consensus (RANSAC) based technique. An iterative nonrigid refinement process using B-spline warping is then used to reconnect axons and produce the sought after connectivity information.


C. Holzhüter, A. Lex, D. Schmalstieg, H. Schulz, H. Schumann, M. Streit.
“Visualizing Uncertainty in Biological Expression Data,” In Proceedings of the SPIE Conference on Visualization and Data Analysis (VDA '12), Vol. 8294, pp. 82940O-82940O-11. 2012.
DOI: 10.1117/12.908516
Expression analysis of ~omics data using microarrays has become a standard procedure in the life sciences. However, microarrays are subject to technical limitations and errors, which render the data gathered likely to be uncertain. While a number of approaches exist to target this uncertainty statistically, it is hardly ever even shown when the data is visualized using for example clustered heatmaps. Yet, this is highly useful when trying not to omit data that is "good enough" for an analysis, which otherwise would be discarded as too unreliable by established conservative thresholds. Our approach addresses this shortcoming by first identifying the margin above the error threshold of uncertain, yet possibly still useful data. It then displays this uncertain data in the context of the valid data by enhancing a clustered heatmap. We employ different visual representations for the different kinds of uncertainty involved. Finally, it lets the user interactively adjust the thresholds, giving visual feedback in the heatmap representation, so that an informed choice on which thresholds to use can be made instead of applying the usual rule-of-thumb cut-offs. We exemplify the usefulness of our concept by giving details for a concrete use case from our partners at the Medical University of Graz, thereby demonstrating our implementation of the general approach.


Y. Hong, S. Joshi, M. Sanchez, M. Styner, M. Niethammer.
“Metamorphic Geodesic Regression,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2012, pp. 197--205. 2012.
We propose a metamorphic geodesic regression approach approximating spatial transformations for image time-series while simultaneously accounting for intensity changes. Such changes occur for example in magnetic resonance imaging (MRI) studies of the developing brain due to myelination. To simplify computations we propose an approximate metamorphic geodesic regression formulation that only requires pairwise computations of image metamorphoses. The approximated solution is an appropriately weighted average of initial momenta. To obtain initial momenta reliably, we develop a shooting method for image metamorphosis.


J. Huang, W. Pei, C. Wen, G. Chen, W. Chen, H. Bao.
“Output-Coherent Image-Space LIC for Surface Flow Visualization,” In Proceedings of the IEEE Pacific Visualization Symposium 2012, Korea, pp. 137--144. 2012.
Image-space line integral convolution (LIC) is a popular approach for visualizing surface vector fields due to its simplicity and high efficiency. To avoid inconsistencies or color blur during the user interactions in the image-space approach, some methods use surface parameterization or 3D volume texture for the effect of smooth transition, which often require expensive computational or memory cost. Furthermore, those methods cannot achieve consistent LIC results in both granularity and color distribution on different scales.
This paper introduces a novel image-space LIC for surface flows that preserves the texture coherence during user interactions. To make the noise textures under different viewpoints coherent, we propose a simple texture mapping technique that is local, robust and effective. Meanwhile, our approach pre-computes a sequence of mipmap noise textures in a coarse-to-fine manner, leading to consistent transition when the model is zoomed. Prior to perform LIC in the image space, the mipmap noise textures are mapped onto each triangle with randomly assigned texture coordinates. Then, a standard image-space LIC based on the projected vector fields is performed to generate the flow texture. The proposed approach is simple and very suitable for GPU acceleration. Our implementation demonstrates consistent and highly efficient LIC visualization on a variety of datasets.


A.H. Huang, B.M. Baker, G.A. Ateshian, R.L. Mauch.
“Sliding Contact Loading Enhances The Tensile Properties Of Mesenchymal Stem Cell-Seeded Hydrogels,” In European Cells and Materials, Vol. 24, pp. 29--45. 2012.
PubMed ID: 22791371
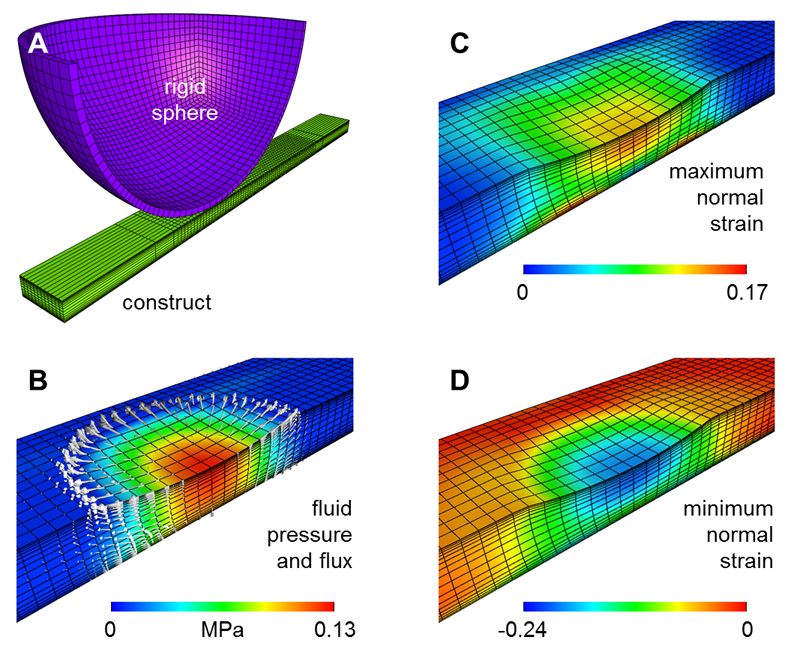
The primary goal of cartilage tissue engineering is to recapitulate the functional properties and structural features of native articular cartilage. While there has been some success in generating near-native compressive properties, the tensile properties of cell-seeded constructs remain poor, and key features of cartilage, including inhomogeneity and anisotropy, are generally absent in these engineered constructs. Therefore, in an attempt to instill these hallmark properties of cartilage in engineered cell-seeded constructs, we designed and characterized a novel sliding contact bioreactor to recapitulate the mechanical stimuli arising from physiologic joint loading (two contacting cartilage layers). Finite element modeling of this bioreactor system showed that tensile strains were direction-dependent, while both tensile strains and fluid motion were depth-dependent and highest in the region closest to the contact surface. Short-term sliding contact of mesenchymal stem cell (MSC)-seeded agarose improved chondrogenic gene expression in a manner dependent on both the axial strain applied and transforming growth factor-? supplementation. Using the optimized loading parameters derived from these short-term studies, long-term sliding contact was applied to MSC-seeded agarose constructs for 21 d. After 21 d, sliding contact significantly improved the tensile properties of MSC-seeded constructs and elicited alterations in type II collagen and proteoglycan accumulation as a function of depth; staining for these matrix molecules showed intense localization in the surface regions. These findings point to the potential of sliding contact to produce engineered cartilage constructs that begin to recapitulate the complex mechanical features of the native tissue.


A. Humphrey, Q. Meng, M. Berzins, T. Harman.
“Radiation Modeling Using the Uintah Heterogeneous CPU/GPU Runtime System,” SCI Technical Report, No. UUSCI-2012-003, SCI Institute, University of Utah, 2012.
The Uintah Computational Framework was developed to provide an environment for solving fluid-structure interaction problems on structured adaptive grids on large-scale, long-running, data-intensive problems. Uintah uses a combination of fluid-flow solvers and particle-based methods for solids, together with a novel asynchronous task-based approach with fully automated load balancing. Uintah demonstrates excellent weak and strong scalability at full machine capacity on XSEDE resources such as Ranger and Kraken, and through the use of a hybrid memory approach based on a combination of MPI and Pthreads, Uintah now runs on up to 262k cores on the DOE Jaguar system. In order to extend Uintah to heterogeneous systems, with ever-increasing CPU core counts and additional onnode GPUs, a new dynamic CPU-GPU task scheduler is designed and evaluated in this study. This new scheduler enables Uintah to fully exploit these architectures with support for asynchronous, outof- order scheduling of both CPU and GPU computational tasks. A new runtime system has also been implemented with an added multi-stage queuing architecture for efficient scheduling of CPU and GPU tasks. This new runtime system automatically handles the details of asynchronous memory copies to and from the GPU and introduces a novel method of pre-fetching and preparing GPU memory prior to GPU task execution. In this study this new design is examined in the context of a developing, hierarchical GPUbased ray tracing radiation transport model that provides Uintah with additional capabilities for heat transfer and electromagnetic wave propagation. The capabilities of this new scheduler design are tested by running at large scale on the modern heterogeneous systems, Keeneland and TitanDev, with up to 360 and 960 GPUs respectively. On these systems, we demonstrate significant speedups per GPU against a standard CPU core for our radiation problem.
Keywords: csafe, uintah


A. Humphrey, Q. Meng, M. Berzins, T. Harman.
“Radiation Modeling Using the Uintah Heterogeneous CPU/GPU Runtime System,” In Proceedings of the first conference of the Extreme Science and Engineering Discovery Environment (XSEDE'12), Association for Computing Machinery, 2012.
DOI: 10.1145/2335755.2335791
The Uintah Computational Framework was developed to provide an environment for solving fluid-structure interaction problems on structured adaptive grids on large-scale, long-running, data-intensive problems. Uintah uses a combination of fluid-flow solvers and particle-based methods for solids, together with a novel asynchronous task-based approach with fully automated load balancing. Uintah demonstrates excellent weak and strong scalability at full machine capacity on XSEDE resources such as Ranger and Kraken, and through the use of a hybrid memory approach based on a combination of MPI and Pthreads, Uintah now runs on up to 262k cores on the DOE Jaguar system. In order to extend Uintah to heterogeneous systems, with ever-increasing CPU core counts and additional onnode GPUs, a new dynamic CPU-GPU task scheduler is designed and evaluated in this study. This new scheduler enables Uintah to fully exploit these architectures with support for asynchronous, outof- order scheduling of both CPU and GPU computational tasks. A new runtime system has also been implemented with an added multi-stage queuing architecture for efficient scheduling of CPU and GPU tasks. This new runtime system automatically handles the details of asynchronous memory copies to and from the GPU and introduces a novel method of pre-fetching and preparing GPU memory prior to GPU task execution. In this study this new design is examined in the context of a developing, hierarchical GPUbased ray tracing radiation transport model that provides Uintah with additional capabilities for heat transfer and electromagnetic wave propagation. The capabilities of this new scheduler design are tested by running at large scale on the modern heterogeneous systems, Keeneland and TitanDev, with up to 360 and 960 GPUs respectively. On these systems, we demonstrate significant speedups per GPU against a standard CPU core for our radiation problem.
Keywords: Uintah, hybrid parallelism, scalability, parallel, adaptive, GPU, heterogeneous systems, Keeneland, TitanDev


A. Irimia, M.C. Chambers, C.M. Torgerson, M. Filippou, D.A. Hovda, J.R. Alger, G. Gerig, A.W. Toga, P.M. Vespa, R. Kikinis, J.D. Van Horn.
“Patient-tailored connectomics visualization for the assessment of white matter atrophy in traumatic brain injury,” In Frontiers in Neurotrauma, Note: http://www.frontiersin.org/neurotrauma/10.3389/fneur.2012.00010/abstract, 2012.
DOI: 10.3389/fneur.2012.00010
Available approaches to the investigation of traumatic brain injury (TBI) are frequently hampered, to some extent, by the unsatisfactory abilities of existing methodologies to efficiently define and represent affected structural connectivity and functional mechanisms underlying TBI-related pathology. In this paper, we describe a patient-tailored framework which allows mapping and characterization of TBI-related structural damage to the brain via multimodal neuroimaging and personalized connectomics. Specifically, we introduce a graphically driven approach for the assessment of trauma-related atrophy of white matter connections between cortical structures, with relevance to the quantification of TBI chronic case evolution. This approach allows one to inform the formulation of graphical neurophysiological and neuropsychological TBI profiles based on the particular structural deficits of the affected patient. In addition, it allows one to relate the findings supplied by our workflow to the existing body of research that focuses on the functional roles of the cortical structures being targeted. Agraphical means for representing patient TBI status is relevant to the emerging field of personalized medicine and to the investigation of neural atrophy.


A. Irimia, Bo Wang, S.R. Aylward, M.W. Prastawa, D.F. Pace, G. Gerig, D.A. Hovda, R.Kikinis, P.M. Vespa, J.D. Van Horn.
“Neuroimaging of Structural Pathology and Connectomics in Traumatic Brain Injury: Toward Personalized Outcome Prediction,” In NeuroImage: Clinical, Vol. 1, No. 1, Elsvier, pp. 1--17. 2012.
DOI: 10.1016/j.nicl.2012.08.002
Recent contributions to the body of knowledge on traumatic brain injury (TBI) favor the view that multimodal neuroimaging using structural and functional magnetic resonance imaging (MRI and fMRI, respectively) as well as diffusion tensor imaging (DTI) has excellent potential to identify novel biomarkers and predictors of TBI outcome. This is particularly the case when such methods are appropriately combined with volumetric/morphometric analysis of brain structures and with the exploration of TBI]related changes in brain network properties at the level of the connectome. In this context, our present review summarizes recent developments on the roles of these two techniques in the search for novel structural neuroimaging biomarkers that have TBI outcome prognostication value. The themes being explored cover notable trends in this area of research, including (1) the role of advanced MRI processing methods in the analysis of structural pathology, (2) the use of brain connectomics and network analysis to identify outcome biomarkers, and (3) the application of multivariate statistics to predict outcome using neuroimaging metrics. The goal of the review is to draw the communityfs attention to these recent advances on TBI outcome prediction methods and to encourage the development of new methodologies whereby structural neuroimaging can be used to identify biomarkers of TBI outcome.


S.K. Iyer, T. Tasdizen, E.V.R. DiBella.
“Edge Enhanced Spatio-Temporal Constrained Reconstruction of Undersampled Dynamic Contrast Enhanced Radial MRI,” In Magnetic Resonance Imaging, Vol. 30, No. 5, pp. 610--619. 2012.
Dynamic contrast-enhanced magnetic resonance imaging (MRI) is a technique used to study and track contrast kinetics in an area of interest in the body over time. Reconstruction of images with high contrast and sharp edges from undersampled data is a challenge. While good results have been reported using a radial acquisition and a spatiotemporal constrained reconstruction (STCR) method, we propose improvements from using spatially adaptive weighting and an additional edge-based constraint. The new method uses intensity gradients from a sliding window reference image to improve the sharpness of edges in the reconstructed image. The method was tested on eight radial cardiac perfusion data sets with 24 rays and compared to the STCR method. The reconstructions showed that the new method, termed edge-enhanced spatiotemporal constrained reconstruction, was able to reconstruct images with sharper edges, and there were a 36\%±13.7\% increase in contrast-to-noise ratio and a 24\%±11\% increase in contrast near the edges when compared to STCR. The novelty of this paper is the combination of spatially adaptive weighting for spatial total variation (TV) constraint along with a gradient matching term to improve the sharpness of edges. The edge map from a reference image allows the reconstruction to trade-off between TV and edge enhancement, depending on the spatially varying weighting provided by the edge map.
Keywords: MRI, Reconstruction, Edge enhanced, Compressed sensing, Regularization, Cardiac perfusion


C.R. Johnson.
“Biomedical Visual Computing: Case Studies and Challenges,” In IEEE Computing in Science and Engineering, Vol. 14, No. 1, pp. 12--21. 2012.
PubMed ID: 22545005
PubMed Central ID: PMC3336198
Computer simulation and visualization are having a substantial impact on biomedicine and other areas of science and engineering. Advanced simulation and data acquisition techniques allow biomedical researchers to investigate increasingly sophisticated biological function and structure. A continuing trend in all computational science and engineering applications is the increasing size of resulting datasets. This trend is also evident in data acquisition, especially in image acquisition in biology and medical image databases.
For example, in a collaboration between neuroscientist Robert Marc and our research team at the University of Utah's Scientific Computing and Imaging (SCI) Institute (www.sci.utah.edu), we're creating datasets of brain electron microscopy (EM) mosaics that are 16 terabytes in size. However, while there's no foreseeable end to the increase in our ability to produce simulation data or record observational data, our ability to use this data in meaningful ways is inhibited by current data analysis capabilities, which already lag far behind. Indeed, as the NIH-NSF Visualization Research Challenges report notes, to effectively understand and make use of the vast amounts of data researchers are producing is one of the greatest scientific challenges of the 21st century.
Visual data analysis involves creating images that convey salient information about underlying data and processes, enabling the detection and validation of expected results while leading to unexpected discoveries in science. This allows for the validation of new theoretical models, provides comparison between models and datasets, enables quantitative and qualitative querying, improves interpretation of data, and facilitates decision making. Scientists can use visual data analysis systems to explore \"what if\" scenarios, define hypotheses, and examine data under multiple perspectives and assumptions. In addition, they can identify connections between numerous attributes and quantitatively assess the reliability of hypotheses. In essence, visual data analysis is an integral part of scientific problem solving and discovery.
As applied to biomedical systems, visualization plays a crucial role in our ability to comprehend large and complex data-data that, in two, three, or more dimensions, convey insight into many diverse biomedical applications, including understanding neural connectivity within the brain, interpreting bioelectric currents within the heart, characterizing white-matter tracts by diffusion tensor imaging, and understanding morphology differences among different genetic mice phenotypes.
Keywords: kaust


E. Jurrus, S. Watanabe, R.J. Giuly, A.R.C. Paiva, M.H. Ellisman, E.M. Jorgensen, T. Tasdizen.
“Semi-Automated Neuron Boundary Detection and Nonbranching Process Segmentation in Electron Microscopy Images,” In Neuroinformatics, pp. (published online). 2012.
Neuroscientists are developing new imaging techniques and generating large volumes of data in an effort to understand the complex structure of the nervous system. The complexity and size of this data makes human interpretation a labor-intensive task. To aid in the analysis, new segmentation techniques for identifying neurons in these feature rich datasets are required. This paper presents a method for neuron boundary detection and nonbranching process segmentation in electron microscopy images and visualizing them in three dimensions. It combines both automated segmentation techniques with a graphical user interface for correction of mistakes in the automated process. The automated process first uses machine learning and image processing techniques to identify neuron membranes that deliniate the cells in each two-dimensional section. To segment nonbranching processes, the cell regions in each two-dimensional section are connected in 3D using correlation of regions between sections. The combination of this method with a graphical user interface specially designed for this purpose, enables users to quickly segment cellular processes in large volumes.


T. Kapur, S. Pieper, R.T. Whitaker, S. Aylward, M. Jakab, W. Schroeder, R. Kikinis.
“The National Alliance for Medical Image Computing, a roadmap initiative to build a free and open source software infrastructure for translational research in medical image analysis,” In Journal of the American Medical Informatics Association, In Journal of the American Medical Informatics Association, Vol. 19, No. 2, pp. 176--180. 2012.
DOI: 10.1136/amiajnl-2011-000493
The National Alliance for Medical Image Computing (NA-MIC), is a multi-institutional, interdisciplinary community of researchers, who share the recognition that modern health care demands improved technologies to ease suffering and prolong productive life. Organized under the National Centers for Biomedical Computing 7 years ago, the mission of NA-MIC is to implement a robust and flexible open-source infrastructure for developing and applying advanced imaging technologies across a range of important biomedical research disciplines. A measure of its success, NA-MIC is now applying this technology to diseases that have immense impact on the duration and quality of life: cancer, heart disease, trauma, and degenerative genetic diseases. The targets of this technology range from group comparisons to subject-specific analysis.


M. Kim, G. Chen, C.D. Hansen.
“Dynamic particle system for mesh extraction on the GPU,” In Proceedings of the 5th Annual Workshop on General Purpose Processing with Graphics Processing Units, London, England, GPGPU-5, ACM, New York, NY, USA pp. 38--46. 2012.
ISBN: 978-1-4503-1233-2
DOI: 10.1145/2159430.215943
Extracting isosurfaces represented as high quality meshes from three-dimensional scalar fields is needed for many important applications, particularly visualization and numerical simulations. One recent advance for extracting high quality meshes for isosurface computation is based on a dynamic particle system. Unfortunately, this state-of-the-art particle placement technique requires a significant amount of time to produce a satisfactory mesh. To address this issue, we study the parallelism property of the particle placement and make use of CUDA, a parallel programming technique on the GPU, to significantly improve the performance of particle placement. This paper describes the curvature dependent sampling method used to extract high quality meshes and describes its implementation using CUDA on the GPU.
Keywords: CUDA, GPGPU, particle systems, volumetric data mesh extraction


J. King, H. Mirzaee, J.K. Ryan, R.M. Kirby.
“Smoothness-Increasing Accuracy-Conserving (SIAC) Filtering for discontinuous Galerkin Solutions: Improved Errors Versus Higher-Order Accuracy,” In Journal of Scientific Computing, Vol. 53, pp. 129--149. 2012.
DOI: 10.1007/s10915-012-9593-8
Smoothness-increasing accuracy-conserving (SIAC) filtering has demonstrated its effectiveness in raising the convergence rate of discontinuous Galerkin solutions from order k + 1/2 to order 2k + 1 for specific types of translation invariant meshes (Cockburn et al. in Math. Comput. 72:577–606, 2003; Curtis et al. in SIAM J. Sci. Comput. 30(1):272– 289, 2007; Mirzaee et al. in SIAM J. Numer. Anal. 49:1899–1920, 2011). Additionally, it improves the weak continuity in the discontinuous Galerkin method to k - 1 continuity. Typically this improvement has a positive impact on the error quantity in the sense that it also reduces the absolute errors. However, not enough emphasis has been placed on the difference between superconvergent accuracy and improved errors. This distinction is particularly important when it comes to understanding the interplay introduced through meshing, between geometry and filtering. The underlying mesh over which the DG solution is built is important because the tool used in SIAC filtering—convolution—is scaled by the geometric mesh size. This heavily contributes to the effectiveness of the post-processor. In this paper, we present a study of this mesh scaling and how it factors into the theoretical errors. To accomplish the large volume of post-processing necessary for this study, commodity streaming multiprocessors were used; we demonstrate for structured meshes up to a 50× speed up in the computational time over traditional CPU implementations of the SIAC filter.


J. Knezevic, R.-P. Mundani, E. Rank, A. Khan, C.R. Johnson.
“Extending the SCIRun Problem Solving Environment to Large-Scale Applications,” In Proceedings of Applied Computing 2012, IADIS, pp. 171--178. October, 2012.
To make the most of current advanced computing technologies, experts in particular areas of science and engineering should be supported by sophisticated tools for carrying out computational experiments. The complexity of individual components of such tools should be hidden from them so they may concentrate on solving the specific problem within their field of expertise. One class of such tools are Problem Solving Environments (PSEs). The contribution of this paper refers to the idea of integration of an interactive computing framework applicable to different engineering applications into the SCIRun PSE in order to enable interactive real-time response of the computational model to user interaction even for large-scale problems. While the SCIRun PSE allows for real-time computational steering, we propose extending this functionality to a wider range of applications and larger scale problems. With only minor code modifications the proposed system allows each module scheduled for execution in a dataflow-based simulation to be automatically interrupted and re-scheduled. This rescheduling allows one to keep the relation between the user interaction and its immediate effect transparent independent of the problem size, thus, allowing for the intuitive and interactive exploration of simulation results.
Keywords: scirun