SCI Publications
2011


J. Mandel, J.D. Beezley, A. Kochanski, V.Y. Kondratenko, L. Zhang, E. Anderson, J. Daniels II, C.T. Silva, C.R. Johnson.
“A wildland fire modeling and visualization environment,” In Proceedings of the Ninth Symposium on Fire and Forest Meteorology, pp. (published online). 2011.


T. Martin, E. Cohen, R.M. Kirby.
“Direct Isosurface Visualization of Hex-Based High-Order Geometry and Attribute Representations,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. PP, No. 99, pp. 1--14. 2011.
ISSN: 1077-2626
DOI: 10.1109/TVCG.2011.103
In this paper, we present a novel isosurface visualization technique that guarantees the accuarate visualization of isosurfaces with complex attribute data defined on (un-)structured (curvi-)linear hexahedral grids. Isosurfaces of high-order hexahedralbased finite element solutions on both uniform grids (including MRI and CT scans) and more complex geometry represent a domain of interest that can be rendered using our algorithm. Additionally, our technique can be used to directly visualize solutions and attributes in isogeometric analysis, an area based on trivariate high-order NURBS (Non-Uniform Rational B-splines) geometry and attribute representations for the analysis. Furthermore, our technique can be used to visualize isosurfaces of algebraic functions. Our approach combines subdivision and numerical root-finding to form a robust and efficient isosurface visualization algorithm that does not miss surface features, while finding all intersections between a view frustum and desired isosurfaces. This allows the use of view-independent transparency in the rendering process. We demonstrate our technique through a straightforward CPU implementation on both complexstructured and complex-unstructured geometry with high-order simulation solutions, isosurfaces of medical data sets, and isosurfaces of algebraic functions.


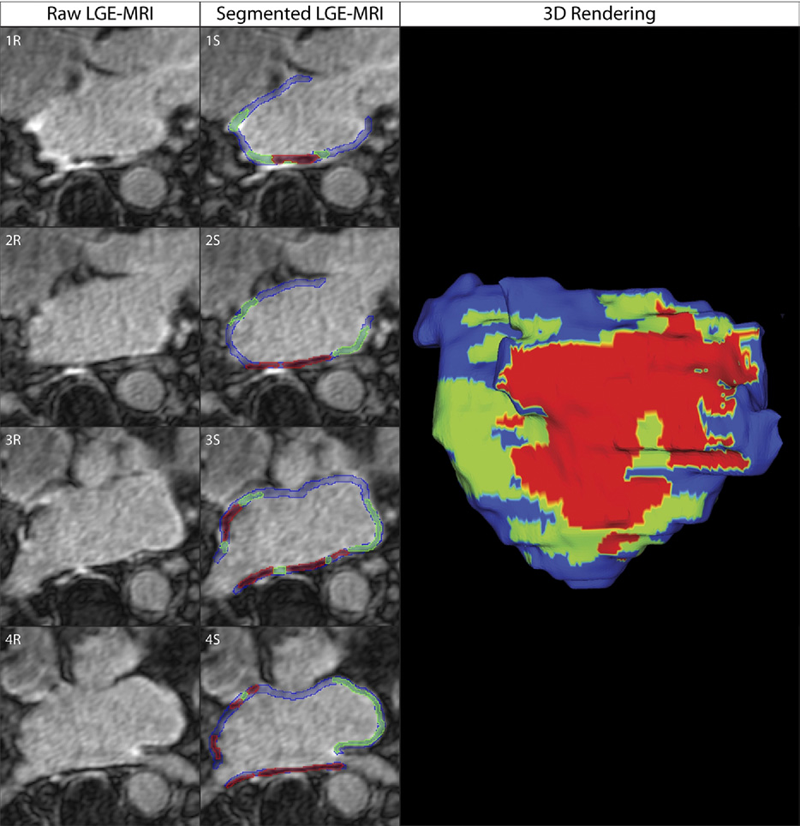
C.J. McGann, E.G. Kholmovski, J.J. Blauer, S. Vijayakumar, T.S. Haslam, J.E. Cates, E.V. DiBella, N.S. Burgon, B. Wilson, A.J. Alexander, M.W. Prastawa, M. Daccarett, G. Vergara, N.W. Akoum, D.L. Parker, R.S. MacLeod, N.F. Marrouche.
“Dark Regions of No-Reflow on Late Gadolinium Enhancement Magnetic Resonance Imaging Result in Scar Formation After Atrial Fibrillation Ablation,” In Journal of the American College of Cardiology, Vol. 58, No. 2, pp. 177--185. 2011.
DOI: 10.1016/j.jacc.2011.04.008
PubMed ID: 21718914
Objectives: The aim of this study was to assess acute ablation injuries seen on late gadolinium enhancement (LGE) magnetic resonance imaging (MRI) immediately post-ablation (IPA) and the association with permanent scar 3 months post-ablation (3moPA).
Background: Success rates for atrial fibrillation catheter ablation vary significantly, in part because of limited information about the location, extent, and permanence of ablation injury at the time of procedure. Although the amount of scar on LGE MRI months after ablation correlates with procedure outcomes, early imaging predictors of scar remain elusive.
Methods: Thirty-seven patients presenting for atrial fibrillation ablation underwent high-resolution MRI with a 3-dimensional LGE sequence before ablation, IPA, and 3moPA using a 3-T scanner. The acute left atrial wall injuries on IPA scans were categorized as hyperenhancing (HE) or nonenhancing (NE) and compared with scar 3moPA.
Results: Heterogeneous injuries with HE and NE regions were identified in all patients. Dark NE regions in the left atrial wall on LGE MRI demonstrate findings similar to the \"no-reflow\" phenomenon. Although the left atrial wall showed similar amounts of HE, NE, and normal tissue IPA (37.7 ± 13\%, 34.3 ± 14\%, and 28.0 ± 11\%, respectively; p = NS), registration of IPA injuries with 3moPA scarring demonstrated that 59.0 ± 19\% of scar resulted from NE tissue, 30.6 ± 15\% from HE tissue, and 10.4 ± 5\% from tissue identified as normal. Paired t-test comparisons were all statistically significant among NE, HE, and normal tissue types (p less than 0.001). Arrhythmia recurrence at 1-year follow-up correlated with the degree of wall enhancement 3moPA (p = 0.02).
Conclusion: Radiofrequency ablation results in heterogeneous injury on LGE MRI with both HE and NE wall lesions. The NE lesions demonstrate no-reflow characteristics and reveal a better predictor of final scar at 3 months. Scar correlates with procedure outcomes, further highlighting the importance of early scar prediction. (J Am Coll Cardiol 2011;58:177–85) © 2011 by the American College of Cardiology Foundation


Q. Meng, M. Berzins, J. Schmidt.
“Using Hybrid Parallelism to improve memory use in Uintah,” In Proceedings of the TeraGrid 2011 Conference, Salt Lake City, Utah, ACM, July, 2011.
DOI: 10.1145/2016741.2016767
The Uintah Software framework was developed to provide an environment for solving fluid-structure interaction problems on structured adaptive grids on large-scale, long-running, data-intensive problems. Uintah uses a combination of fluid-flow solvers and particle-based methods for solids together with a novel asynchronous task-based approach with fully automated load balancing. Uintah's memory use associated with ghost cells and global meta-data has become a barrier to scalability beyond O(100K) cores. A hybrid memory approach that addresses this issue is described and evaluated. The new approach based on a combination of Pthreads and MPI is shown to greatly reduce memory usage as predicted by a simple theoretical model, with comparable CPU performance.
Keywords: Uintah, C-SAFE, parallel computing


H. Mirzaee, Liangyue, J.K. Ryan, R.M. Kirby.
“Smoothness-Increasing Accuracy-Conserving (SIAC) Postprocessing for Discontinuous Galerkin Solutions Over Structured Triangular Meshes,” In SIAM Journal of Numerical Analysis, Vol. 49, No. 5, pp. 1899--1920. 2011.
Theoretically and computationally, it is possible to demonstrate that the order of accuracy of a discontinuous Galerkin (DG) solution for linear hyperbolic equations can be improved from order k+1 to 2k+1 through the use of smoothness-increasing accuracy-conserving (SIAC) filtering. However, it is a computationally complex task to perform this in an efficient manner, which becomes an even greater issue considering nonquadrilateral mesh structures. In this paper, we present an extension of this SIAC filter to structured triangular meshes. The basic theoretical assumption in the previous implementations of the postprocessor limits the use to numerical solutions solved over a quadrilateral mesh. However, this assumption is restrictive, which in turn complicates the application of this postprocessing technique to general tessellations. Additionally, moving from quadrilateral meshes to triangulated ones introduces more complexity in the calculations as the number of integrations required increases. In this paper, we extend the current theoretical results to variable coefficient hyperbolic equations over structured triangular meshes and demonstrate the effectiveness of the application of this postprocessor to structured triangular meshes as well as exploring the effect of using inexact quadrature. We show that there is a direct theoretical extension to structured triangular meshes for hyperbolic equations with bounded variable coefficients. This is a challenging first step toward implementing SIAC filters for unstructured tessellations. We show that by using the usual B-spline implementation, we are able to improve on the order of accuracy as well as decrease the magnitude of the errors. These results are valid regardless of whether exact or inexact integration is used. The results here demonstrate that it is still possible, both theoretically and computationally, to improve to 2k+1 over the DG solution itself for structured triangular meshes.


H. Mirzaee, J.K. Ryan, R.M. Kirby.
“Efficient Implementation of Smoothness-Increasing Accuracy-Conserving (SIAC) Filters for Discontinuous Galerkin Solutions,” In Journal of Scientific Computing, pp. (in press). 2011.
DOI: 10.1007/s10915-011-9535-x
The discontinuous Galerkin (DG) methods provide a high-order extension of the finite volume method in much the same way as high-order or spectral/hp elements extend standard finite elements. However, lack of inter-element continuity is often contrary to the smoothness assumptions upon which many post-processing algorithms such as those used in visualization are based. Smoothness-increasing accuracy-conserving (SIAC) filters were proposed as a means of ameliorating the challenges introduced by the lack of regularity at element interfaces by eliminating the discontinuity between elements in a way that is consistent with the DG methodology; in particular, high-order accuracy is preserved and in many cases increased. The goal of this paper is to explicitly define the steps to efficient computation of this filtering technique as applied to both structured triangular and quadrilateral meshes. Furthermore, as the SIAC filter is a good candidate for parallelization, we provide, for the first time, results that confirm anticipated performance scaling when parallelized on a shared-memory multi-processor machine.


M.J. Mlodzianoski, J.M. Schreiner, S.P. Callahan, K. Smolková, A. Dlasková, J. Šantorová, P. Ježek, J. Bewersdorf.
“Sample drift correction in 3D fluorescence photoactivation localization microscopy,” In Optics Express, Vol. 19, No. 16, pp. 15009--15019. 2011.
DOI: 10.1364/OE.19.015009



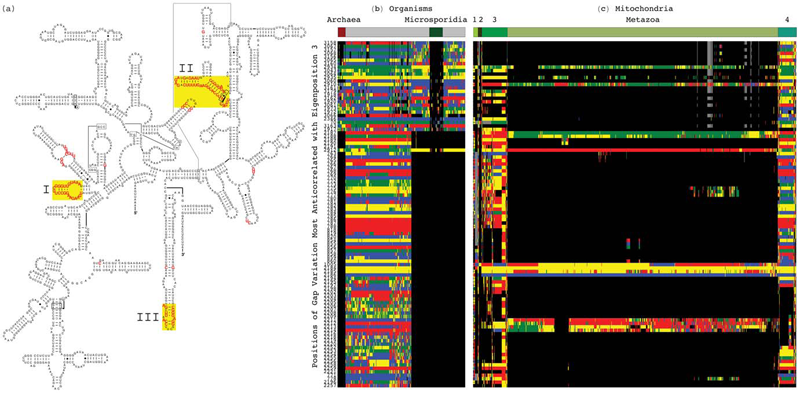
C. Muralidhara, A.M. Gross, R.R. Gutell, O. Alter.
“Tensor Decomposition Reveals Concurrent Evolutionary Convergences and Divergences and Correlations with Structural Motifs in Ribosomal RNA,” In PLoS ONE, Vol. 6, No. 4, Public Library of Science, pp. e18768. April, 2011.
DOI: 10.1371/journal.pone.0018768
Evolutionary relationships among organisms are commonly described by using a hierarchy derived from comparisons of ribosomal RNA (rRNA) sequences. We propose that even on the level of a single rRNA molecule, an organism's evolution is composed of multiple pathways due to concurrent forces that act independently upon different rRNA degrees of freedom. Relationships among organisms are then compositions of coexisting pathway-dependent similarities and dissimilarities, which cannot be described by a single hierarchy. We computationally test this hypothesis in comparative analyses of 16S and 23S rRNA sequence alignments by using a tensor decomposition, i.e., a framework for modeling composite data. Each alignment is encoded in a cuboid, i.e., a third-order tensor, where nucleotides, positions and organisms, each represent a degree of freedom. A tensor mode-1 higher-order singular value decomposition (HOSVD) is formulated such that it separates each cuboid into combinations of patterns of nucleotide frequency variation across organisms and positions, i.e., \"eigenpositions\" and corresponding nucleotide-specific segments of \"eigenorganisms,\" respectively, independent of a-priori knowledge of the taxonomic groups or rRNA structures. We find, in support of our hypothesis that, first, the significant eigenpositions reveal multiple similarities and dissimilarities among the taxonomic groups. Second, the corresponding eigenorganisms identify insertions or deletions of nucleotides exclusively conserved within the corresponding groups, that map out entire substructures and are enriched in adenosines, unpaired in the rRNA secondary structure, that participate in tertiary structure interactions. This demonstrates that structural motifs involved in rRNA folding and function are evolutionary degrees of freedom. Third, two previously unknown coexisting subgenic relationships between Microsporidia and Archaea are revealed in both the 16S and 23S rRNA alignments, a convergence and a divergence, conferred by insertions and deletions of these motifs, which cannot be described by a single hierarchy. This shows that mode-1 HOSVD modeling of rRNA alignments might be used to computationally predict evolutionary mechanisms.


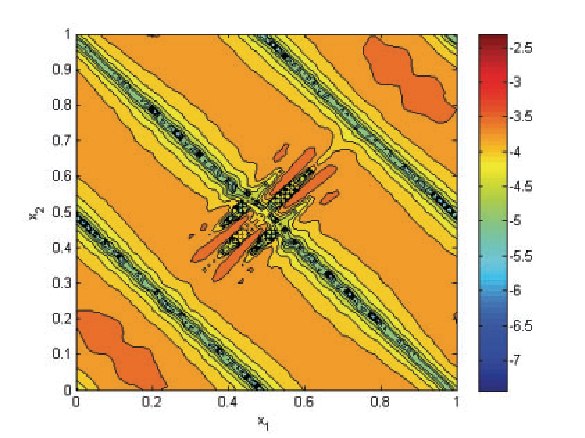
A. Narayan, D. Xiu.
“Distributional Sensitivity for Uncertainty Quantification,” In Communications in Computational Physics, Vol. 10, No. 1, pp. 140--160. 2011.
DOI: 10.4208/cicp.160210.300710a
In this work we consider a general notion of distributional sensitivity, which measures the variation in solutions of a given physical/mathematical system with respect to the variation of probability distribution of the inputs. This is distinctively different from the classical sensitivity analysis, which studies the changes of solutions with respect to the values of the inputs. The general idea is measurement of sensitivity of outputs with respect to probability distributions, which is a well-studied concept in related disciplines. We adapt these ideas to present a quantitative framework in the context of uncertainty quantification for measuring such a kind of sensitivity and a set of efficient algorithms to approximate the distributional sensitivity numerically. A remarkable feature of the algorithms is that they do not incur additional computational effort in addition to a one-time stochastic solver. Therefore, an accurate stochastic computation with respect to a prior input distribution is needed only once, and the ensuing distributional sensitivity computation for different input distributions is a post-processing step. We prove that an accurate numericalmodel leads to accurate calculations of this sensitivity, which applies not just to slowly-converging Monte-Carlo estimates, but also to exponentially convergent spectral approximations. We provide computational examples to demonstrate the ease of applicability and verify the convergence claims.
Keywords: Uncertainty quantification, epistemic uncertainty, distributional sensitivity, generalized polynomial chaos


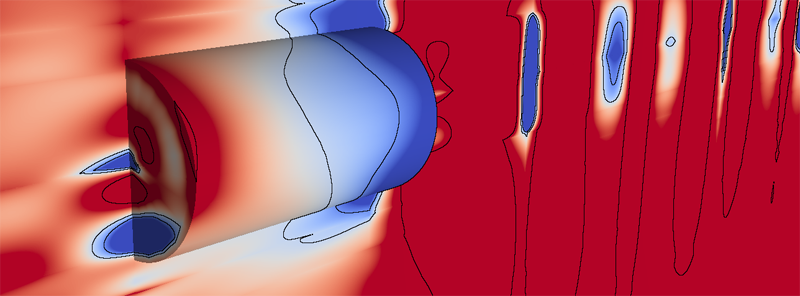
B. Nelson, R. Haimes, R.M. Kirby.
“GPU-Based Interactive Cut-Surface Extraction From High-0rder Finite Element Fields,” In IEEE Transactions on Visualization and Computer Graphics (IEEE Visualization Issue), Vol. 17, No. 12, pp. 1803--1811. 2011.
We present a GPU-based ray-tracing system for the accurate and interactive visualization of cut-surfaces through 3D simulations of physical processes created from spectral/hp high-order finite element methods. When used by the numerical analyst to debug the solver, the ability for the imagery to precisely reflect the data is critical. In practice, the investigator interactively selects from a palette of visualization tools to construct a scene that can answer a query of the data. This is effective as long as the implicit contract of image quality between the individual and the visualization system is upheld. OpenGL rendering of scientific visualizations has worked remarkably well for exploratory visualization for most solver results. This is due to the consistency between the use of first-order representations in the simulation and the linear assumptions inherent in OpenGL (planar fragments and color-space interpolation). Unfortunately, the contract is broken when the solver discretization is of higher-order. There have been attempts to mitigate this through the use of spatial adaptation and/or texture mapping. These methods do a better job of approximating what the imagery should be but are not exact and tend to be view-dependent. This paper introduces new rendering mechanisms that specifically deal with the kinds of native data generated by high-order finite element solvers. The exploratory visualization tools are reassessed and cast in this system with the focus on image accuracy. This is accomplished in a GPU setting to ensure interactivity.


J.T. Oden, O. Ghattas, J.L. King, B.I. Schneider, K. Bartschat, F. Darema, J. Drake, T. Dunning, D. Estep, S. Glotzer, M. Gurnis, C.R. Johnson, D.S. Katz, D. Keyes, S. Kiesler, S. Kim, J. Kinter, G. Klimeck, C.W. McCurdy, R. Moser, C. Ott, A. Patra, L. Petzold, T. Schlick, K. Schulten, V. Stodden, J. Tromp, M. Wheeler, S.J. Winter, C. Wu, K. Yelick.
“Cyber Science and Engineering: A Report of the National Science Foundation Advisory Committee for Cyberinfrastructure Task Force on Grand Challenges,” Note: NSF Report, 2011.
This document contains the findings and recommendations of the NSF – Advisory Committee for Cyberinfrastructure Task Force on Grand Challenges addressed by advances in Cyber Science and Engineering. The term Cyber Science and Engineering (CS&E) is introduced to describe the intellectual discipline that brings together core areas of science and engineering, computer science, and computational and applied mathematics in a concerted effort to use the cyberinfrastructure (CI) for scientific discovery and engineering innovations; CS&E is computational and data-based science and engineering enabled by CI. The report examines a host of broad issues faced in addressing the Grand Challenges of science and technology and explores how those can be met by advances in CI. Included in the report are recommendations for new programs and initiatives that will expand the portfolio of the Office of Cyberinfrastructure and that will be critical to advances in all areas of science and engineering that rely on the CI.


Y. Pan, W.-K. Jeong, R.T. Whitaker.
“Markov surfaces: A probabilistic framework for user-assisted three-dimensional image segmentation,” In Computer Vision and Image Understanding, Vol. 115, No. 10, pp. 1375--1383. 2011.
This paper presents Markov surfaces, a probabilistic algorithm for user-assisted segmentation of elongated structures in 3D images. The 3D segmentation problem is formulated as a path-finding problem, where path probabilities are described by Markov chains. Users define points, curves, or regions on 2D image slices, and the algorithm connects these user-defined features in a way that respects the underlying elongated structure in data. Transition probabilities in the Markov model are derived from intensity matches and interslice correspondences, which are generated from a slice-to-slice registration algorithm. Bezier interpolations between paths are applied to generate smooth surfaces. Subgrid accuracy is achieved by linear interpolations of image intensities and the interslice correspondences. Experimental results on synthetic and real data demonstrate that Markov surfaces can segment regions that are defined by texture, nearby context, and motion. A parallel implementation on a streaming parallel computer architecture, a graphics processor, makes the method interactive for 3D data.

Valerio Pascucci, Xavier Tricoche, Hans Hagen, Julien Tierny.
“Topological Methods in Data Analysis and Visualization: Theory, Algorithms, and Applications (Mathematics and Visualization),” Springer, 2011.
ISBN: 978-3642150135


T. Peterka, R. Ross, A. Gyulassy, V. Pascucci, W. Kendall, H.-W. Shen, T.-Y. Lee, A. Chaudhuri.
“Scalable Parallel Building Blocks for Custom Data Analysis,” In Proceedings of the 2011 IEEE Symposium on Large-Scale Data Analysis and Visualization (LDAV), pp. 105--112. October, 2011.
DOI: 10.1109/LDAV.2011.6092324
We present a set of building blocks that provide scalable data movement capability to computational scientists and visualization researchers for writing their own parallel analysis. The set includes scalable tools for domain decomposition, process assignment, parallel I/O, global reduction, and local neighborhood communicationtasks that are common across many analysis applications. The global reduction is performed with a new algorithm, described in this paper, that efficiently merges blocks of analysis results into a smaller number of larger blocks. The merging is configurable in the number of blocks that are reduced in each round, the number of rounds, and the total number of resulting blocks. We highlight the use of our library in two analysis applications: parallel streamline generation and parallel Morse-Smale topological analysis. The first case uses an existing local neighborhood communication algorithm, whereas the latter uses the new merge algorithm.


S. Philip, B. Summa, P.-T. Bremer, and V. Pascucci.
“Parallel Gradient Domain Processing of Massive Images,” In Proceedings of the 2011 Eurographics Symposium on Parallel Graphics and Visualization, pp. 11--19. 2011.
Gradient domain processing remains a particularly computationally expensive technique even for relatively small images. When images become massive in size, giga or terapixel, these problems become particularly troublesome and the best serial techniques take on the order of hours or days to compute a solution. In this paper, we provide a simple framework for the parallel gradient domain processing. Specifically, we provide a parallel out-of-core method for the seamless stitching of gigapixel panoramas in a parallel MPI environment. Unlike existing techniques, the framework provides both a straightforward implementation, maintains strict control over the required/allocated resources, and makes no assumptions on the speed of convergence to an acceptable image. Furthermore, the approach shows good weak/strong scaling from several to hundreds of cores and runs on a variety of hardware.


S. Philip, B. Summa, P-T Bremer, V. Pascucci.
“Hybrid CPU-GPU Solver for Gradient Domain Processing of Massive Images,” In Proceedings of 2011 International Conference on Parallel and Distributed Systems (ICPADS), pp. 244--251. 2011.
Gradient domain processing is a computationally expensive image processing technique. Its use for processing massive images, giga or terapixels in size, can take several hours with serial techniques. To address this challenge, parallel algorithms are being developed to make this class of techniques applicable to the largest images available with running times that are more acceptable to the users. To this end we target the most ubiquitous form of computing power available today, which is small or medium scale clusters of commodity hardware. Such clusters are continuously increasing in scale, not only in the number of nodes, but also in the amount of parallelism available within each node in the form of multicore CPUs and GPUs. In this paper we present a hybrid parallel implementation of gradient domain processing for seamless stitching of gigapixel panoramas that utilizes MPI, threading and a CUDA based GPU component. We demonstrate the performance and scalability of our implementation by presenting results from two GPU clusters processing two large data sets.


T.A. Quinn, S. Granite, M.A. Allessie, C. Antzelevitch, C. Bollensdorff, G. Bub, R.A.B. Burton, E. Cerbai, P.S. Chen, M. Delmar, D. DiFrancesco, Y.E. Earm, I.R. Efimov, M. Egger, E. Entcheva, M. Fink, R. Fischmeister, M.R. Franz, A. Garny, W.R. Giles, T. Hannes, S.E. Harding, P.J. Hunter, s, G. Iribe, J. Jalife, C.R. Johnson, R.S. Kass, I. Kodama, G. Koren, P. Lord, V.S. Markhasin, S. Matsuoka, A.D. McCulloch, G.R. Mirams, G.E. Morley, S. Nattel, D. Noble, S.P. Olesen, A.V. Panfilov, N.A. Trayanova, U. Ravens, S. Richard, D.S. Rosenbaum, Y. Rudy, F. Sachs, F.B. Sachse, D.A. Saint, U. Schotten, O. Solovyova, P. Taggart, L. Tung, A. Varrò, P.G. Volders, K. Wang, J.N. Weiss, E. Wettwer, E. White, R. Wilders, R.L. Winslow, P. Kohl.
“Minimum Information about a Cardiac Electrophysiology Experiment (MICEE): Standardised reporting for model reproducibility, interoperability, and data sharing,” In Progress in Biophysics and Molecular Biology, Vol. 107, No. 1, Elsevier, pp. 4--10. October, 2011.
DOI: 10.1016/j.pbiomolbio.2011.07.001
PubMed Central ID: PMC3190048
Cardiac experimental electrophysiology is in need of a well-defined Minimum Information Standard for recording, annotating, and reporting experimental data. As a step toward establishing this, we present a draft standard, called Minimum Information about a Cardiac Electrophysiology Experiment (MICEE). The ultimate goal is to develop a useful tool for cardiac electrophysiologists which facilitates and improves dissemination of the minimum information necessary for reproduction of cardiac electrophysiology research, allowing for easier comparison and utilisation of findings by others. It is hoped that this will enhance the integration of individual results into experimental, computational, and conceptual models. In its present form, this draft is intended for assessment and development by the research community. We invite the reader to join this effort, and, if deemed productive, implement the Minimum Information about a Cardiac Electrophysiology Experiment standard in their own work.
Keywords: Minimum Information Standard; Cardiac electrophysiology; Data sharing; Reproducibility; Integration; Computational modelling


W. Reich, Dominic Schneider, Christian Heine, Alexander Wiebel, Guoning Chen, Gerik Scheuermann.
“Combinatorial Vector Field Topology in 3 Dimensions,” In Mathematical Methods in Biomedical Image Analysis (MMBIA) Proceedings IEEE MMBIA 2012, pp. 47--59. November, 2011.
DOI: 10.1007/978-3-642-23175-9_4
In this paper, we present two combinatorial methods to process 3-D steady vector fields, which both use graph algorithms to extract features from the underlying vector field. Combinatorial approaches are known to be less sensitive to noise than extracting individual trajectories. Both of the methods are a straightforward extension of an existing 2-D technique to 3-D fields. We observed that the first technique can generate overly coarse results and therefore we present a second method that works using the same concepts but produces more detailed results. We evaluate our method on a CFD-simulation of a gas furnace chamber. Finally, we discuss several possibilities for categorizing the invariant sets with respect to the flow.


P. Rosen, V. Popescu, K. Hayward, C. Wyman.
“Non-Pinhole Approximations for Interactive Rendering,” In IEEE Computer Graphics and Applications, Vol. 99, 2011.


P. Rosen, V. Popescu.
“An Evaluation of 3-D Scene Exploration Using a Multiperspective Image Framework,” In The Visual Computer, Vol. 27, No. 6-8, Springer-Verlag New York, Inc., pp. 623--632. 2011.
DOI: 10.1007/s00371-011-0599-2
PubMed ID: 22661796
PubMed Central ID: PMC3364594
Multiperspective images (MPIs) show more than what is visible from a single viewpoint and are a promising approach for alleviating the problem of occlusions. We present a comprehensive user study that investigates the effectiveness of MPIs for 3-D scene exploration. A total of 47 subjects performed searching, counting, and spatial orientation tasks using both conventional and multiperspective images. We use a flexible MPI framework that allows trading off disocclusion power for image simplicity. The framework also allows rendering MPI images at interactive rates, which enables investigating interactive navigation and dynamic 3-D scenes. The results of our experiments show that MPIs can greatly outperform conventional images. For searching, subjects performed on average 28% faster using an MPI. For counting, accuracy was on average 91% using MPIs as compared to 42% for conventional images.
Keywords: Interactive 3-D scene exploration, Navigation, Occlusions, User study, Visual interfaces