SCI Publications
2009


N.M. Segerson, M. Daccarett, T.J. Badger, A. Shabaan, N. Akoum, E.N. Fish, S. Rao, N.S. Burgon, Y. Adjei-Poku, E.G. Kholmovski, S. Vijayakumar, E.V.R. Dibella, R.S. Macleod, N.F. Marrouche.
“Magnetic Resonance Imaging-Confirmed Ablative Debulking of the Left Atrial Posterior Wall and Septum for Treatment of Persistent Atrial Fibrillation: Rationale and Initial Experience,” In Journal of Cardiovascular Electrophysiology, Vol. 21, No. 2, pp. 126--132. 2009.


J.F. Shepherd, C.R. Johnson.
“Hexahedral Mesh Generation for Biomedical Models in SCIRun,” In Engineering with Computers, Vol. 25, No. 1, pp. 97--114. 2009.


F. Shi, P.T. Yap, Y. Fan, J.Z. Cheng, L.L. Wald, G. Gerig, W. Lin, D. Shen.
“Cortical Enhanced Tissue Segmentation of Neonatal Brain MR Images Acquired by a Dedicated Phased Array Coil,” In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 39--45. 2009.
PubMed ID: 20862268


M. Steffen.
“Analysis-Guided Improvements of the Material Point Method,” School of Computing, University of Utah, 2009.
The Material Point Method (MPM) has shown itself to be a powerful tool in the simulation of large deformation problems, especially those involving complex geometries and contact where typical finite element type methods frequently fail. While these large complex problems lead to some impressive simulations and solutions, there has been a lack of basic analysis characterizing the errors present in the method, even on the simplest of problems. However, like most methods which employ mixed Lagrangian (particle) and Eulerian strategies, analysis of the method is not straight forward. The lack of an analysis framework for MPM, as is found in finite element methods, makes it challenging to explain anomalies found in its employment and makes it difficult to propose methodology improvements with predictable outcomes. In this dissertation, we provide a formal analysis of the errors in MPM and use this analysis to direct proposed improvements. In particular, we will focus on how the lack of regularity in the grid functions used for representing the solution can hamper both spatial and temporal convergence of the method. We will show how the use of smoother basis functions, such as B-splines, can improve the accuracy of the method. An in-depth analysis of the current time stepping methods will help to explain behavior currently demonstrated numerically in the literature and will allow users of the method to understand the balance of spatial and temporal errors in MPM. Lastly, extrapolation techniques will be proposed to improve quadrature errors in the method.


M. Streit, A. Lex, H. Müller, D. Schmalstieg.
“Gaze-Based Focus Adaption in an Information Visualization System,” In Proceedings of the Conference on Computer Graphics and Visualization and Image Processing (CGVCVIP '09), 2009.
As the complexity and amount of real world data continuously grows, modern visualization systems are changing. Traditional information visualization techniques are often not sufficient to allow an in-depth visual data exploration process. Multiple view systems combined with linking & brushing are only one building block of a successful InfoVis system. In this paper we propose the incorporation of cheap and simple gaze-based interaction. We employ the tracking information not for selecting data (i.e. mouse interaction) but for an intelligent adaption of 2D and 3D visualizations. Derived from the focus+context paradigm, we call this gaze-focus. The proposed methods are demonstrated by means of three different visualizations.


M. Streit, A. Lex, M. Kalkusch, K. Zatloukal, D. Schmalstieg.
“Caleydo: Connecting Pathways with Gene Expression,” In Bioinformatics, Vol. 25, No. 20, pp. 2760--2761. 2009.
Understanding the relationships between pathways and the altered expression of their components in disease conditions can be addressed in a visual data analysis process. Caleydo uses novel visualization techniques to support life science experts in their analysis of gene expression data in the context of pathways and functions of individual genes. Pathways and gene expression visualizations are placed in a 3D scene where selected entities (i.e. genes) are visually connected. This allows Caleydo to seamlessly integrate interactive gene expression visualization with cross-database pathway exploration.


R. Tao, P.T. Fletcher, R.T. Whitaker.
“A Variational Image-Based Approach to the Correction of Susceptibility Artifacts in the Alignment of Diffusion Weighted and Structural MRI,” In Lecture Notes in Computer Science, Springer, pp. 664--675. 2009.
DOI: 10.1007/978-3-642-02498-6_55
PubMed ID: 19694302


T. Tasdizen.
“Principal neighborhood dictionaries for nonlocal means image denoising,” In IEEE Transactions on Image Processing, Vol. 18, No. 12, Note: Epub 2009 Jul 24, pp. 2649--2660. 2009.
PubMed ID: 19635697


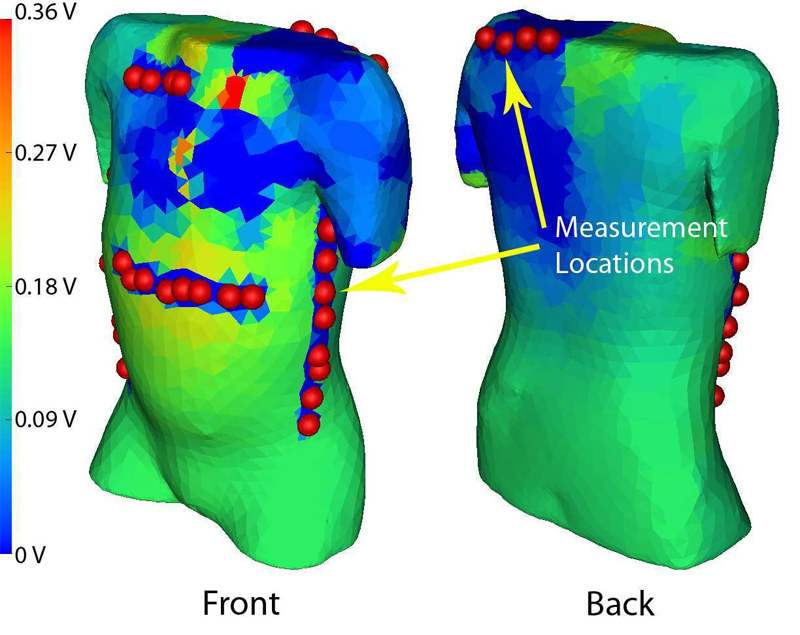
J.D. Tate, J.G. Stinstra, T. Pilcher, and R.S. MacLeod.
“Measuring Implantable Cardioverter Defibrillators (ICDs) During Implantation Surgery: Verification of a Simulation,” In Computers in Cardiology, pp. 473--476. 2009.
ISSN: 0276-6547
Implantable cardioverter defibrillators (ICDs) are increasing used in abnormal configurations. We have developed a patient specific forward simulation model to predict efficacy of the defibrillation shock. Our goal was to develop a method of measuring the ICD surface potentials as the devices are tested during implantation surgery to use as verification of the simulation. A lead selection algorithm was used to develop a surface potential mapping system with 32 recording sites that do not interfere with implantation surgery. ICD discharge recordings were compared at similar locations to corresponding patient models. The reconstructed simulated surface potentials showed


J. Tierny, A. Gyulassy, E. Simon, V. Pascucci.
“Loop Surgery for Volumetric Meshes: Reeb Graphs Reduced to Contour Trees,” In IEEE Transactions on Visualization and Computer Graphics, Proceedings of the 2009 IEEE Visualization Conference, Vol. 15, No. 6, pp. 1177--1184. Sept/Oct, 2009.
DOI: 10.1109/TVCG.2009.163


L.T. Tran, J. Kim, M. Berzins.
“Solving Time-Dependent PDEs using the Material Point Method, A Case Study from Gas Dynamics,” In International Journal for Numerical Methods in Fluids, Vol. 62, No. 7, pp. 709--732. 2009.


Kannan U.V., A.R.C. Paiva, E. Jurrus, T. Tasdizen.
“Automatic Markup of Neural Cell Membranes Using Boosted Decision Stumps,” In Proceedings of the IEEE International Symposium on Biomedical Engineering (ISBI 2009), Boston, MA, pp. 1039--1042. 2009.
DOI: 10.1109/ISBI.2009.5193233
To better understand the central nervous system, neurobiologists need to reconstruct the underlying neural circuitry from electron microscopy images. One of the necessary tasks is to segment the individual neurons. For this purpose, we propose a supervised learning approach to detect the cell membranes. The classifier was trained using AdaBoost, on local and context features. The features were selected to highlight the line characteristics of cell membranes. It is shown that using features from context positions allows for more information to be utilized in the classification. Together with the nonlinear discrimination ability of the AdaBoost classifier, this results in clearly noticeable improvements over previously used methods.
Keywords: crcns, neural networks


Kannan UV, M. Kim, D. Gerszewski, J.R. Anderson, M. Hall.
“Assembling Large Mosaics of Electron Microscope Images using GPU,” In Proceedings of the 2009 Symposium on Application Accelerators in High Performance Computing (SAAHPC'09), 2009.
DOI: 10.1.1.163.213
Understanding the neural circuitry of the retina requires us to map the connectivity of individual neurons in large neuronal tissue sections and analyze signal communication across processes from the electron microscopy images. One of the major bottlenecks in the critical path is the image mosaicing process where 2D slices are assembled from scanned microscopy image tiles. The problem of assembling the tiles is computationally non-trivial because of distortion of the specimen in the electron microscope due to heat and overlap between the scanned tiles. The complexity of the calculation arises from the massive size of the dataset and mathematical calculations required to calculate value of each pixel of the mosaic. We propose to use texture memory lookups to speedup the access to image tiles and data parallel computing enabled by the GPUs to accelerate this process. The proposed method results in noticeable improvements in speed of computation compared to other methods. Index Terms—Serial-section TEM, image mosaicing, GPGPU, CUDA, texture.
Keywords: crcns, electron microscopy, mosaics, retina, eye


A. Vo, S. Vakkalanka, M. Delisi, G. Gopalakrishnan, R.M. Kirby, R. Thakur.
“Formal Verification of Practical MPI Programs,” In Proceedings of 14th ACM SIGPLAN Symposium on Principles and Practice of Parallel Programming (PPoPP), Raleigh, NC, pp. 261--270. February 14-18, 2009.


H.T. Vo, D.K. Osmari, B. Summa, J.L.D. Comba, V. Pascucci, C.T. Silva.
“Parallel Dataflow Scheme for Streaming (Un)Structured Data,” SCI Technical Report, No. UUSCI-2009-004, SCI Institute, University of Utah, 2009.


H.T. Vo, C.T. Silva.
“Multi-Threaded Streaming Pipeline For VTK,” SCI Technical Report, No. UUSCI-2009-005, SCI Institute, University of Utah, 2009.


I. Wald, W.R. Mark, J. Günther, S. Boulos, T. Ize, W. Hunt, S.G. Parker, P. Shirley.
“State of the Art in Ray Tracing Animated Scenes,” In Computer Graphics Forum, Vol. 28, No. 6, pp. 1691--1722. 2009.
DOI: 10.1111/j.1467-8659.2008.01313.x


M. Waldner, A. Lex, M. Streit, D. Schmalstieg.
“Design Considerations for Collaborative Information Workspaces in Multi-Display Environments,” In Proceedings of the Workshop on Collaborative Visualization on Interactive Surfaces (VisWeek '09), pp. 36--39. 2009.
ISSN: 1862-5207
The incorporation of massive amounts of data from different sources is a challenging task for the conception of any information visualization system. Especially the data heterogeneity often makes it necessary to include people from multiple domains with various fields of expertise. Hence, the inclusion of multiple users in a collaborative data analysis process introduces a whole new challenge for the design and conception of visualization applications. Using a multi-display environment to support co-located collaborative work seems to be a natural next step. However, adapting common visualization systems to multi-display environments poses several challenges.
We have come up with a number of design considerations for employing multiple-view visualizations in collaborative multi-display environments: adaptations of the visualization depending on display factors and user preferences, interaction techniques to facilitate information sharing and to guide the users' attention to relevant items in the environment, and the design of a flexible working environment, adjustable to varying group sizes and specific tasks.
Motivated by these considerations we propose a system relying on a spatial model of the environment as its main information source. We argue that the system design should be separated into basic multi-display environment functionality, such as multiple input handling and the management of the physical displays, and higher level functionality provided by the visualization system. An API offered by the multi-display framework thereby provides the necessary information about the environment and users to the visualization system.


Y. Wan, H. Otsuna, C.-B. Chien, C.D. Hansen.
“An Interactive Visualization Tool for Multi-channel Confocal Microscopy Data in Neurobiology Research,” SCI Technical Report, No. UUSCI-2009-001, SCI Institute, University of Utah, 2009.


Y. Wan, H. Otsuna, C.-B. Chien, C.D. Hansen.
“An Interactive Visualization Tool for Multi-Channel Confocal Microscopy Data in Neurobiology Research,” In IEEE Transactions on Visualization and Computer Graphics, Proceedings of the 2009 IEEE Visualization Conference, Vol. 15, No. 6, pp. 1489--1496. Sept/Oct, 2009.