SCI Publications
2011


R.C. Knickmeyer, C. Kang, S. Woolson, K.J. Smith, R.M. Hamer, W. Lin, G. Gerig, M. Styner, J.H. Gilmore.
“Twin-Singleton Differences in Neonatal Brain Structure,” In Twin Research and Human Genetics, Vol. 14, No. 3, pp. 268--276. 2011.
ISSN: 1832-4274
DOI: 10.1375/twin.14.3.268


A. Knoll, S. Thelen, I. Wald, C.D. Hansen, H. Hagen, M.E. Papka.
“Full-Resolution Interactive CPU Volume Rendering with Coherent BVH Traversal,” In Proceedings of IEEE Pacific Visualization 2011, pp. 3--10. 2011.


B.H. Kopell, J. Halverson, C.R. Butson, M. Dickinson, J. Bobholz, H. Harsch, C. Rainey, D. Kondziolka, R. Howland, E. Eskandar, K.C. Evans, D.D. Dougherty.
“Epidural cortical stimulation of the left dorsolateral prefrontal cortex for refractory major depressive disorder,” In Neurosurgery, Vol. 69, No. 5, pp. 1015--1029. November, 2011.
ISSN: 1524-4040
DOI: 10.1227/NEU.0b013e318229cfcd
A significant number of patients with major depressive disorder are unresponsive to conventional therapies. For these patients, neuromodulation approaches are being investigated.


S. Kumar, V. Vishwanath, P. Carns, B. Summa, G. Scorzelli, V. Pascucci, R. Ross, J. Chen, H. Kolla, R. Grout.
“PIDX: Efficient Parallel I/O for Multi-resolution Multi-dimensional Scientific Datasets,” In Proceedings of The IEEE International Conference on Cluster Computing, pp. 103--111. September, 2011.


S. Kurugol, E. Bas, D. Erdogmus, J.G. Dy, G.C. Sharp, D.H. Brooks.
“Centerline extraction with principal curve tracing to improve 3D level set esophagus segmentation in CT images,” In Proceedings of IEEE International Conference of the Engineering in Medicine and Biology Society (EMBS), pp. 3403--3406. 2011.
DOI: 10.1109/IEMBS.2011.6090921
PubMed ID: 2225507
PubMed Central ID: PMC3349355
For radiotherapy planning, contouring of target volume and healthy structures at risk in CT volumes is essential. To automate this process, one of the available segmentation techniques can be used for many thoracic organs except the esophagus, which is very hard to segment due to low contrast. In this work we propose to initialize our previously introduced model based 3D level set esophagus segmentation method with a principal curve tracing (PCT) algorithm, which we adapted to solve the esophagus centerline detection problem. To address challenges due to low intensity contrast, we enhanced the PCT algorithm by learning spatial and intensity priors from a small set of annotated CT volumes. To locate the esophageal wall, the model based 3D level set algorithm including a shape model that represents the variance of esophagus wall around the estimated centerline is utilized. Our results show improvement in esophagus segmentation when initialized by PCT compared to our previous work, where an ad hoc centerline initialization was performed. Unlike previous approaches, this work does not need a very large set of annotated training images and has similar performance.


S. Kurugol, J.G. Dy, M. Rajadhyaksha, K.W. Gossage, J. Weissman, D.H. Brooks.
“Semi-automated Algorithm for Localization of Dermal/ Epidermal Junction in Reflectance Confocal Microscopy Images of Human Skin,” In Proceedings of SPIE, Vol. 7904, pp. 79041A-79041A-10. 2011.
DOI: 10.1117/12.875392
PubMed ID: 21709746
PubMed Central ID: PMC3120112
The examination of the dermis/epidermis junction (DEJ) is clinically important for skin cancer diagnosis. Reflectance confocal microscopy (RCM) is an emerging tool for detection of skin cancers in vivo. However, visual localization of the DEJ in RCM images, with high accuracy and repeatability, is challenging, especially in fair skin, due to low contrast, heterogeneous structure and high inter- and intra-subject variability. We recently proposed a semi-automated algorithm to localize the DEJ in z-stacks of RCM images of fair skin, based on feature segmentation and classification. Here we extend the algorithm to dark skin. The extended algorithm first decides the skin type and then applies the appropriate DEJ localization method. In dark skin, strong backscatter from the pigment melanin causes the basal cells above the DEJ to appear with high contrast. To locate those high contrast regions, the algorithm operates on small tiles (regions) and finds the peaks of the smoothed average intensity depth profile of each tile. However, for some tiles, due to heterogeneity, multiple peaks in the depth profile exist and the strongest peak might not be the basal layer peak. To select the correct peak, basal cells are represented with a vector of texture features. The peak with most similar features to this feature vector is selected. The results show that the algorithm detected the skin types correctly for all 17 stacks tested (8 fair, 9 dark). The DEJ detection algorithm achieved an average distance from the ground truth DEJ surface of around 4.7μm for dark skin and around 7-14μm for fair skin.
Keywords: confocal reflectance microscopy, image analysis, skin, classification


S. Kurugol, J.G. Dy, D.H. Brooks, M. Rajadhyaksha.
“Pilot study of semiautomated localization of the dermal/epidermal junction in reflectance confocal microscopy images of skin,” In Journal of biomedical optics, Vol. 16, No. 3, International Society for Optics and Photonics, pp. 036005--036005. 2011.
DOI: 10.1117/1.3549740
Reflectance confocal microscopy (RCM) continues to be translated toward the detection of skin cancers in vivo. Automated image analysis may help clinicians and accelerate clinical acceptance of RCM. For screening and diagnosis of cancer, the dermal/epidermal junction (DEJ), at which melanomas and basal cell carcinomas originate, is an important feature in skin. In RCM images, the DEJ is marked by optically subtle changes and features and is difficult to detect purely by visual examination. Challenges for automation of DEJ detection include heterogeneity of skin tissue, high inter-, intra-subject variability, and low optical contrast. To cope with these challenges, we propose a semiautomated hybrid sequence segmentation/classification algorithm that partitions z-stacks of tiles into homogeneous segments by fitting a model of skin layer dynamics and then classifies tile segments as epidermis, dermis, or transitional DEJ region using texture features. We evaluate two different training scenarios: 1. training and testing on portions of the same stack; 2. training on one labeled stack and testing on one from a different subject with similar skin type. Initial results demonstrate the detectability of the DEJ in both scenarios with epidermis/dermis misclassification rates smaller than 10% and average distance from the expert labeled boundaries around 8.5 μm.


M. Leeser, D. Yablonski, D.H. Brooks, L.S. King.
“The Challenges of Writing Portable, Correct and High Performance Libraries for GPUs,” In Computer Architecture News, Vol. 39, No. 4, pp. 2--7. 2011.
DOI: 10.1145/2082156.2082158
Graphics Processing Units (GPUs) are widely used to accelerate scientific applications. Many successes have been reported with speedups of two or three orders of magnitude over serial implementations of the same algorithms. These speedups typically pertain to a specific implementation with fixed parameters mapped to a specific hardware implementation. The implementations are not designed to be easily ported to other GPUs, even from the same manufacturer. When target hardware changes, the application must be re-optimized.
In this paper we address a different problem. We aim to deliver working, efficient GPU code in a library that is downloaded and run by many different users. The issue is to deliver efficiency independent of the individual user parameters and without a priori knowledge of the hardware the user will employ. This problem requires a different set of tradeoffs than finding the best runtime for a single solution. Solutions must be adaptable to a range of different parameters both to solve users' problems and to make the best use of the target hardware.
Another issue is the integration of GPUs into a Problem Solving Environment (PSE) where the use of a GPU is almost invisible from the perspective of the user. Ease of use and smooth interactions with the existing user interface are important to our approach. We illustrate our solution with the incorporation of GPU processing into the Scientific Computing Institute (SCI)Run Biomedical PSE developed at the University of Utah. SCIRun allows scientists to interactively construct many different types of biomedical simulations. We use this environment to demonstrate the effectiveness of the GPU by accelerating time consuming algorithms in the scientist's simulations. Specifically we target the linear solver module, including Conjugate Gradient, Jacobi and MinRes solvers for sparse matrices.


Z. Leng, J.R. Korenberg, B. Roysam, T. Tasdizen.
“A rapid 2-D centerline extraction method based on tensor voting,” In 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 1000--1003. 2011.
DOI: 10.1109/ISBI.2011.5872570


A. Lex, H. Schulz, M. Streit, C. Partl, D. Schmalstieg.
“VisBricks: Multiform Visualization of Large, Inhomogeneous Data,” In IEEE Transactions on Visualization and Computer Graphics (InfoVis '11), Vol. 17, No. 12, 2011.
Large volumes of real-world data often exhibit inhomogeneities: vertically in the form of correlated or independent dimensions and horizontally in the form of clustered or scattered data items. In essence, these inhomogeneities form the patterns in the data that researchers are trying to find and understand. Sophisticated statistical methods are available to reveal these patterns, however, the visualization of their outcomes is mostly still performed in a one-view-fits-all manner, In contrast, our novel visualization approach, VisBricks, acknowledges the inhomogeneity of the data and the need for different visualizations that suit the individual characteristics of the different data subsets. The overall visualization of the entire data set is patched together from smaller visualizations, there is one VisBrick for each cluster in each group of interdependent dimensions. Whereas the total impression of all VisBricks together gives a comprehensive high-level overview of the different groups of data, each VisBrick independently shows the details of the group of data it represents, State-of-the-art brushing and visual linking between all VisBricks furthermore allows the comparison of the groupings and the distribution of data items among them. In this paper, we introduce the VisBricks visualization concept, discuss its design rationale and implementation, and demonstrate its usefulness by applying it to a use case from the field of biomedicine.


G. Li, R. Palmer, M. DeLisi, G. Gopalakrishnan, R.M. Kirby.
“Formal Specification of MPI 2.0: Case Study in Specifying a Practical Concurrent Programming API,” In Science of Computer Programming, Vol. 76, pp. 65--81. 2011.
DOI: 10.1016/j.scico.2010.03.007
We describe the first formal specification of a non-trivial subset of MPI, the dominant communication API in high performance computing. Engineering a formal specification for a non-trivial concurrency API requires the right combination of rigor, executability, and traceability, while also serving as a smooth elaboration of a pre-existing informal specification. It also requires the modularization of reusable specification components to keep the length of the specification in check. Long-lived APIs such as MPI are not usually 'textbook minimalistic' because they support a diverse array of applications, a diverse community of users, and have efficient implementations over decades of computing hardware. We choose the TLA+ notation to write our specifications, and describe how we organized the specification of around 200 of the 300 MPI 2.0 functions. We detail a handful of these functions in this paper, and assess our specification with respect to the aforementioned requirements. We close with a description of possible approaches that may help render the act of writing, understanding, and validating the specifications of concurrency APIs much more productive.


J. Li, J. Li, D. Xiu.
“An Efficient Surrogate-based Method for Computing Rare Failure Probability,” In Journal of Computational Physics, Vol. 230, No. 24, pp. 8683--8697. 2011.
DOI: 10.1016/j.jcp.2011.08.008
In this paper, we present an efficient numerical method for evaluating rare failure probability. The method is based on a recently developed surrogate-based method from Li and Xiu [J. Li, D. Xiu, Evaluation of failure probability via surrogate models, J. Comput. Phys. 229 (2010) 8966–8980] for failure probability computation. The method by Li and Xiu is of hybrid nature, in the sense that samples of both the surrogate model and the true physical model are used, and its efficiency gain relies on using only very few samples of the true model. Here we extend the capability of the method to rare probability computation by using the idea of importance sampling (IS). In particular, we employ cross-entropy (CE) method, which is an effective method to determine the biasing distribution in IS. We demonstrate that, by combining with the CE method, a surrogate-based IS algorithm can be constructed and is highly efficient for rare failure probability computation—it incurs much reduced simulation efforts compared to the traditional CE-IS method. In many cases, the new method is capable of capturing failure probability as small as 10-12 ~ 10-6 with only several hundreds samples.
Keywords: Rare events, Failure probability, Importance sampling, Cross-entropy


L. Lins, D. Koop, J. Freire, C.T. Silva.
“DEFOG: A System for Data-Backed Visual Composition,” SCI Technical Report, No. UUSCI-2011-003, SCI Institute, University of Utah, 2011.


W. Liu, S. Awate, J. Anderson, D. Yurgelun-Todd, P.T. Fletcher.
“Monte Carlo expectation maximization with hidden Markov models to detect functional networks in resting-state fMRI,” In Machine Learning in Medical Imaging, Lecture Notes in Computer Science (LNCS), Vol. 7009/2011, pp. 59--66. 2011.
DOI: 10.1007/978-3-642-24319-6_8


J. Luitjens, M. Berzins.
“Scalable parallel regridding algorithms for block-structured adaptive mesh renement,” In Concurrency And Computation: Practice And Experience, Vol. 23, No. 13, John Wiley & Sons, Ltd., pp. 1522--1537. 2011.
ISSN: 1532--0634
DOI: 10.1002/cpe.1719


J.P. Luitjens.
“The Scalability of Parallel Adaptive Mesh Refinement Within Uintah,” Note: Advisor: Martin Berzins, School of Computing, University of Utah, 2011.
Solutions to Partial Differential Equations (PDEs) are often computed by discretizing the domain into a collection of computational elements referred to as a mesh. This solution is an approximation with an error that decreases as the mesh spacing decreases. However, decreasing the mesh spacing also increases the computational requirements. Adaptive mesh refinement (AMR) attempts to reduce the error while limiting the increase in computational requirements by refining the mesh locally in regions of the domain that have large error while maintaining a coarse mesh in other portions of the domain. This approach often provides a solution that is as accurate as that obtained from a much larger fixed mesh simulation, thus saving on both computational time and memory. However, historically, these AMR operations often limit the overall scalability of the application.
Adapting the mesh at runtime necessitates scalable regridding and load balancing algorithms. This dissertation analyzes the performance bottlenecks for a widely used regridding algorithm and presents two new algorithms which exhibit ideal scalability. In addition, a scalable space-filling curve generation algorithm for dynamic load balancing is also presented. The performance of these algorithms is analyzed by determining their theoretical complexity, deriving performance models, and comparing the observed performance to those performance models. The models are then used to predict performance on larger numbers of processors. This analysis demonstrates the necessity of these algorithms at larger numbers of processors. This dissertation also investigates methods to more accurately predict workloads based on measurements taken at runtime. While the methods used are not new, the application of these methods to the load balancing process is. These methods are shown to be highly accurate and able to predict the workload within 3% error. By improving the accuracy of these estimations, the load imbalance of the simulation can be reduced, thereby increasing the overall performance.


J. Luitjens, M. Berzins.
“Scalable parallel regridding algorithms for block-structured adaptive mesh refinement,” In Concurrency and Computation: Practice and Experience, Vol. 23, No. 13, pp. 1522--1537. September, 2011.
DOI: 10.1002/cpe.1719
Block-structured adaptive mesh refinement (BSAMR) is widely used within simulation software because it improves the utilization of computing resources by refining the mesh only where necessary. For BSAMR to scale onto existing petascale and eventually exascale computers all portions of the simulation need to weak scale ideally. Any portions of the simulation that do not will become a bottleneck at larger numbers of cores. The challenge is to design algorithms that will make it possible to avoid these bottlenecks on exascale computers. One step of existing BSAMR algorithms involves determining where to create new patches of refinement. The Berger–Rigoutsos algorithm is commonly used to perform this task. This paper provides a detailed analysis of the performance of two existing parallel implementations of the Berger– Rigoutsos algorithm and develops a new parallel implementation of the Berger–Rigoutsos algorithm and a tiled algorithm that exhibits ideal scalability. The analysis and computational results up to 98 304 cores are used to design performance models which are then used to predict how these algorithms will perform on 100 M cores.


S.A. Maas, B.J. Ellis, D.S. Rawlins, L.T. Edgar, C.R. Henak, J.A. Weiss.
“Implementation and Verification of a Nodally-Integrated Tetrahedral Element in FEBio,” SCI Technical Report, No. UUSCI-2011-007, SCI Institute, University of Utah, 2011.
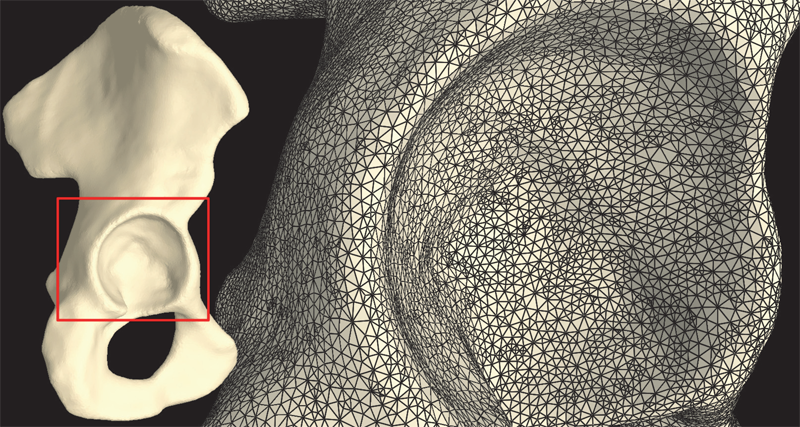
Finite element simulations in computational biomechanics commonly require the discretization of extremely complicated geometries. Creating meshes for these complex geometries can be very difficult and time consuming using hexahedral elements. Automatic meshing algorithms exist for tetrahedral elements, but these elements often have numerical problems that discourage their use in complex finite element models. To overcome these problems we have implemented a stabilized, nodally-integrated tetrahedral element formulation in FEBio, our in-house developed finite element code, allowing researchers to use linear tetrahedral elements in their models and still obtain accurate solutions. In addition to facilitating automatic mesh generation, this also allows researchers to use mesh refinement algorithms which are fairly well developed for tetrahedral elements but not so much for hexahedral elements. In this document, the implementation of the stabilized, nodallyintegrated, tetrahedral element, named the \"UT4 element\", is described. Two slightly different variations of the nodally integrated tetrahedral element are considered. In one variation the entire virtual work is stabilized and in the other one the stabilization is only applied to the isochoric part of the virtual work. The implementation of both formulations has been verified and the convergence behavior illustrated using the patch test and three verification problems. Also, a model from our laboratory with very complex geometry is discretized and analyzed using the UT4 element to show its utility for a problem from the biomechanics literature. The convergence behavior of the UT4 element does vary depending on problem, tetrahedral mesh structure and choice of formulation parameters, but the results from the verification problems should assure analysts that a converged solution using the UT4 element can be obtained that is more accurate than the solution from a classical linear tetrahedral formulation.
Keywords: MRL


R.S. MacLeod, J.J.E. Blauer.
“Atrial Fibrillation,” In Multimodal Cardiovascular Imaging: Principles and Clinical Applications, Ch. 25, Edited by O. Pahlm and G. Wagner, McGraw Hill, 2011.
ISBN: 0071613463
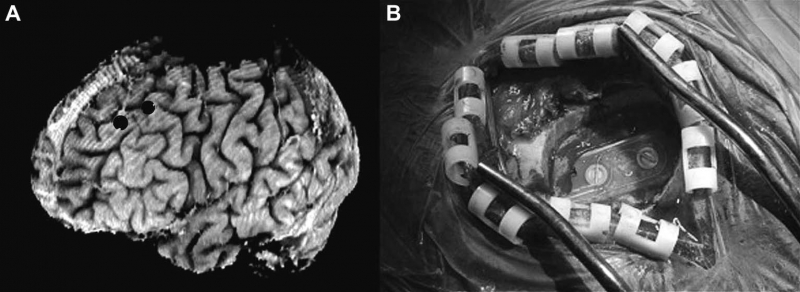
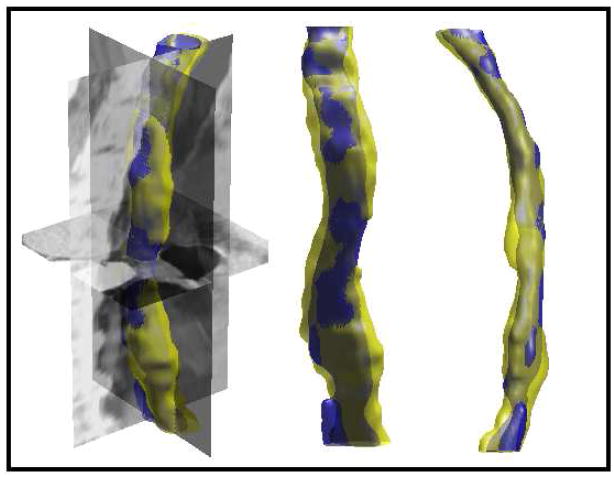
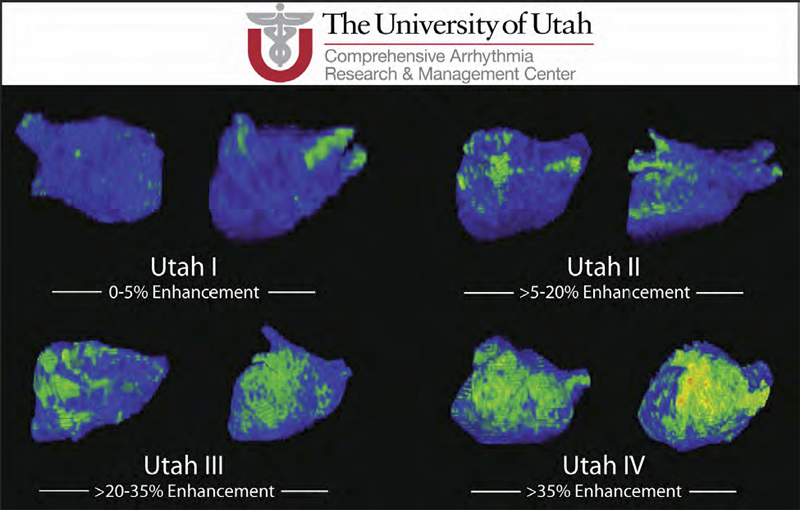
Atrial fibrillation (AF) is the most common form of cardiac arrhythmia so that a review of the role imaging in AF is a natural topic to include in this book. Further motivation comes from the fact that the treatment of AF probably includes more different forms of imaging, often merged or combined in a variety of ways, than perhaps any other clinical intervention. A typical clinical electrophysiology lab for the treatment of AF usually contains no less than 6 and often more than 8 individual monitors, each rendering some form of image based information about the patient undergoing therapy. There is naturally great motivation to merge different images and different imaging modalities in the setting of AF but also very challenging because of a host of factors related to the small size, extremely thin walls, the large natural variation in atrial shape, and the fact that fibrillation is occurring so that atrial shape is changing rapidly and irregularly. Thus, the use of multimodal imaging has recently become a very active and challenging area of image processing and analysis research and development, driven by an enormous clinical need to understand and treat a disease that affects some 5 million Americans alone, a number that is predicted to increase to almost 16 million by 2050.
In this chapter we attempt to provide an overview of the large variety of imaging modalities and uses in the management and understanding of atrial fibrillation, with special emphasis on the most novel applications of magnetic resonance imaging (MRI) technology. To provide clinical and biomedical motivation, we outline the basics of the disease together with some contemporary hypotheses about its etiology and management. We then describe briefly the imaging modalities in common use in the management and research of AF, then focus on the use or MRI for all phases of the management of patients with AF and indicate some of the major engineering challenges that can motivate further progress.
Keywords: ablation, carma, cvrti, 5P41-RR012553-10


M.Q. Madrigal, G. Tadmor, G. C. Cano, D.H. Brooks.
“Low-Order 4D Dynamical Modeling of Heart Motion Under Respiration,” In Proceeding of the IEEE International Symposium on Biomedical Imaging: from nano to macro, pp. 1326--1329. 2011.
PubMed ID: 21927642
PubMed Central ID: PMC3172964
This work is motivated by the limitations of current techniques to visualize the heart as it moves under contraction and respiration during interventional procedures such as ablation of atrial fibrillation. Our long-term goal is to integrate high resolution models routinely obtained from pre-procedure imaging (here, via MRI) with the low resolution, sparse, images, along with a few scalar measurements such as ECG, which are feasible during the real-time procedure. A key ingredient to facilitate this integration is the extraction from the pre-procedure model of an individualized, low complexity, dynamic model of the moving and beating heart. This is the immediate goal we address here. Our approach stems from work on distributed parameter dynamical systems and uses a combination of truncated basis expansions to obtain the requisite four dimensional low order model. The method's potential is illustrated not only by modeling results but also by estimation of an arbitrary slice from the parameterized model.