SCI Publications
2014


B.R. Parmar, T.R. Jarrett, N.S. Burgon, E.G. Kholmovski, N.W. Akoum, N. Hu, R.S. Macleod, N.F. Marrouche, R. Ranjan.
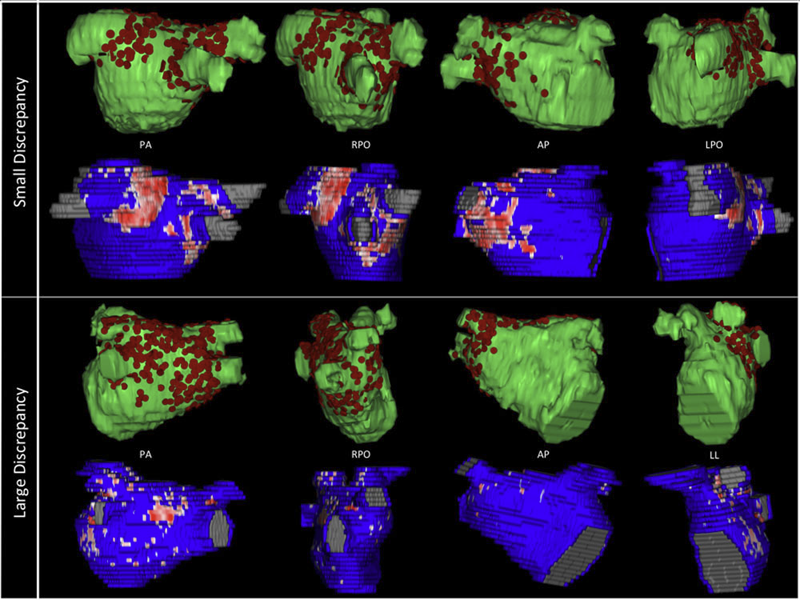
“Comparison of Left Atrial Area Marked Ablated in Electroanatomical Maps with Scar in MRI,” In Journal of Cardiovascular Electrophysiology, 2014.
DOI: 10.1111/jce.12357
Background
Three-dimensional electroanatomic mapping (EAM) is routinely used to mark ablated areas during radiofrequency ablation. We hypothesized that, in atrial fibrillation (AF) ablation, EAM overestimates scar formation in the left atrium (LA) when compared to the scar seen on late-gadolinium enhancement magnetic resonance imaging (LGE-MRI).
Methods and Results
Of the 235 patients who underwent initial ablation for AF at our institution between August 2011 and December 2012, we retrospectively identified 70 patients who had preprocedural magnetic resonance angiography merged with LA anatomy in EAM software and had a 3-month postablation LGE-MRI for assessment of scar. Ablated area was marked intraprocedurally using EAM software and quantified retrospectively. Scarred area was quantified in 3-month postablation LGE-MRI. The mean ablated area in EAM was 30.5 ± 7.5% of the LA endocardial surface and the mean scarred area in LGE-MRI was 13.9 ± 5.9% (P < 0.001). This significant difference in the ablated area marked in the EAM and scar area in the LGE-MRI was present for each of the 3 independent operators. Complete pulmonary vein (PV) encirclement representing electrical isolation was observed in 87.8% of the PVs in EAM as compared to only 37.4% in LGE-MRI (P < 0.001).
Conclusions
In AF ablation, EAM significantly overestimates the resultant scar as assessed with a follow-up LGE-MRI.
Keywords: atrial fibrillation, magnetic resonance imaging, radiofrequency ablation


Christian Partl, Alexander Lex, Marc Streit, Hendrik Strobelt, Anne-Mai Wasserman, Hanspeter Pfister,, Dieter Schmalstieg.
“ConTour: Data-Driven Exploration of Multi-Relational Datasets for Drug Discovery,” In IEEE Transactions on Visualization and Computer Graphics (VAST '14), Vol. 20, No. 12, pp. 1883--1892. 2014.
ISSN: 1077-2626
DOI: 10.1109/TVCG.2014.2346752
Large scale data analysis is nowadays a crucial part of drug discovery. Biologists and chemists need to quickly explore and evaluate potentially effective yet safe compounds based on many datasets that are in relationship with each other. However, there is a lack of tools that support them in these processes. To remedy this, we developed ConTour, an interactive visual analytics technique that enables the exploration of these complex, multi-relational datasets. At its core ConTour lists all items of each dataset in a column. Relationships between the columns are revealed through interaction: selecting one or multiple items in one column highlights and re-sorts the items in other columns. Filters based on relationships enable drilling down into the large data space. To identify interesting items in the first place, ConTour employs advanced sorting strategies, including strategies based on connectivity strength and uniqueness, as well as sorting based on item attributes. ConTour also introduces interactive nesting of columns, a powerful method to show the related items of a child column for each item in the parent column. Within the columns, ConTour shows rich attribute data about the items as well as information about the connection strengths to other datasets. Finally, ConTour provides a number of detail views, which can show items from multiple datasets and their associated data at the same time. We demonstrate the utility of our system in case studies conducted with a team of chemical biologists, who investigate the effects of chemical compounds on cells and need to understand the underlying mechanisms.

A. J. Perez, M. Seyedhosseini, T. J. Deerinck, E. A. Bushong, S. Panda, T. Tasdizen, M. H. Ellisman.
“A workflow for the automatic segmentation of organelles in electron microscopy image stacks,” In Frontiers in Neuroanatomy, Vol. 8, No. 126, 2014.
DOI: 10.3389/fnana.2014.00126
Electron microscopy (EM) facilitates analysis of the form, distribution, and functional status of key organelle systems in various pathological processes, including those associated with neurodegenerative disease. Such EM data often provide important new insights into the underlying disease mechanisms. The development of more accurate and efficient methods to quantify changes in subcellular microanatomy has already proven key to understanding the pathogenesis of Parkinson's and Alzheimer's diseases, as well as glaucoma. While our ability to acquire large volumes of 3D EM data is progressing rapidly, more advanced analysis tools are needed to assist in measuring precise three-dimensional morphologies of organelles within data sets that can include hundreds to thousands of whole cells. Although new imaging instrument throughputs can exceed teravoxels of data per day, image segmentation and analysis remain significant bottlenecks to achieving quantitative descriptions of whole cell structural organellomes. Here, we present a novel method for the automatic segmentation of organelles in 3D EM image stacks. Segmentations are generated using only 2D image information, making the method suitable for anisotropic imaging techniques such as serial block-face scanning electron microscopy (SBEM). Additionally, no assumptions about 3D organelle morphology are made, ensuring the method can be easily expanded to any number of structurally and functionally diverse organelles. Following the presentation of our algorithm, we validate its performance by assessing the segmentation accuracy of different organelle targets in an example SBEM dataset and demonstrate that it can be efficiently parallelized on supercomputing resources, resulting in a dramatic reduction in runtime.

A. Perez, M. Seyedhosseini, T. Tasdizen, M. Ellisman.
“Automated workflows for the morphological characterization of organelles in electron microscopy image stacks (LB72),” In The FASEB Journal, Vol. 28, No. 1 Supplement LB72, April, 2014.
Advances in three-dimensional electron microscopy (EM) have facilitated the collection of image stacks with a field-of-view that is large enough to cover a significant percentage of anatomical subdivisions at nano-resolution. When coupled with enhanced staining protocols, such techniques produce data that can be mined to establish the morphologies of all organelles across hundreds of whole cells in their in situ environments. Although instrument throughputs are approaching terabytes of data per day, image segmentation and analysis remain significant bottlenecks in achieving quantitative descriptions of whole cell organellomes. Here we describe computational workflows that achieve the automatic segmentation of organelles from regions of the central nervous system by applying supervised machine learning algorithms to slices of serial block-face scanning EM (SBEM) datasets. We also demonstrate that our workflows can be parallelized on supercomputing resources, resulting in a dramatic reduction of their run times. These methods significantly expedite the development of anatomical models at the subcellular scale and facilitate the study of how these models may be perturbed following pathological insults.


N. Ramesh, T. Tasdizen.
“Cell tracking using particle filters with implicit convex shape model in 4D confocal microscopy images,” In 2014 IEEE International Conference on Image Processing (ICIP), IEEE, Oct, 2014.
DOI: 10.1109/icip.2014.7025089
Bayesian frameworks are commonly used in tracking algorithms. An important example is the particle filter, where a stochastic motion model describes the evolution of the state, and the observation model relates the noisy measurements to the state. Particle filters have been used to track the lineage of cells. Propagating the shape model of the cell through the particle filter is beneficial for tracking. We approximate arbitrary shapes of cells with a novel implicit convex function. The importance sampling step of the particle filter is defined using the cost associated with fitting our implicit convex shape model to the observations. Our technique is capable of tracking the lineage of cells for nonmitotic stages. We validate our algorithm by tracking the lineage of retinal and lens cells in zebrafish embryos.


F. Rousset, C. Vachet, C. Conlin, M. Heilbrun, J.L. Zhang, V.S. Lee, G. Gerig.
“Semi-automated application for kidney motion correction and filtration analysis in MR renography,” In Proceeding of the 2014 Joint Annual Meeting ISMRM-ESMRMB, pp. (accepted). 2014.
Altered renal function commonly affects patients with cirrhosis, a consequence of chronic liver disease. From lowdose contrast material-enhanced magnetic resonance (MR) renography, we can estimate the Glomerular Filtration Rate (GFR), an important parameter to assess renal function. Two-dimensional MR images are acquired every 2 seconds for approximately 5 minutes during free breathing, which results in a dynamic series of 140 images representing kidney filtration over time. This specific acquisition presents dynamic contrast changes but is also challenged by organ motion due to breathing. Rather than use conventional image registration techniques, we opted for an alternative method based on object detection. We developed a novel analysis framework available under a stand-alone toolkit to efficiently register dynamic kidney series, manually select regions of interest, visualize the concentration curves for these ROIs, and fit them into a model to obtain GFR values. This open-source cross-platform application is written in C++, using the Insight Segmentation and Registration Toolkit (ITK) library, and QT4 as a graphical user interface.


N. Sadeghi, J.H. Gilmore, W. Lin, G. Gerig.
“Normative Modeling of Early Brain Maturation from Longitudinal DTI Reveals Twin-Singleton Differences,” In Proceeding of the 2014 Joint Annual Meeting ISMRM-ESMRMB, pp. (accepted). 2014.
Early brain development of white matter is characterized by rapid organization and structuring. Magnetic Resonance diffusion tensor imaging (MR-DTI) provides the possibility of capturing these changes non-invasively by following individuals longitudinally in order to better understand departures from normal brain development in subjects at risk for mental illness [1]. Longitudinal imaging of individuals suggests the use of 4D (3D, time) image analysis and longitudinal statistical modeling [3].


N. Sadeghi, P.T. Fletcher, M. Prastawa, J.H. Gilmore, G. Gerig.
“Subject-specific prediction using nonlinear population modeling: Application to early brain maturation from DTI,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention (MICCAI 2014), 2014.
The term prediction implies expected outcome in the future, often based on a model and statistical inference. Longitudinal imaging studies offer the possibility to model temporal change trajectories of anatomy across populations of subjects. In the spirit of subject-specific analysis, such normative models can then be used to compare data from new subjects to the norm and to study progression of disease or to predict outcome. This paper follows a statistical inference approach and presents a framework for prediction of future observations based on past measurements and population statistics. We describe prediction in the context of nonlinear mixed effects modeling (NLME) where the full reference population's statistics (estimated fixed effects, variance-covariance of random effects, variance of noise) is used along with the individual's available observations to predict its trajectory. The proposed methodology is generic in regard to application domains. Here, we demonstrate analysis of early infant brain maturation from longitudinal DTI with up to three time points. Growth as observed in DTI-derived scalar invariants is modeled with a parametric function, its parameters being input to NLME population modeling. Trajectories of new subject's data are estimated when using no observation, only the rst or the first two time points. Leave-one-out experiments result in statistics on differences between actual and predicted observations. We also simulate a clinical scenario of prediction on multiple categories, where trajectories predicted from multiple models are classified based on maximum likelihood criteria.


A.R. Sanderson.
“An Alternative Formulation of Lyapunov Exponents for Computing Lagrangian Coherent Structures,” In Proceedings of the 2014 IEEE Pacific Visualization Symposium (PacificVis), Yokahama Japan, 2014.
Lagrangian coherent structures are time-evolving surfaces that highlight areas in flow fields where neighboring advected particles diverge or converge. The detection and understanding of such structures is an important part of many applications such as in oceanography where there is a need to predict the dispersion of oil and other materials in the ocean. One of the most widely used tools for revealing Lagrangian coherent structures has been to calculate the finite-time Lyapunov exponents, whose maximal values appear as ridgelines to reveal Lagrangian coherent structures. In this paper we explore an alternative formulation of Lyapunov exponents for computing Lagrangian coherent structures.

M. Seyedhosseini, T. Tasdizen.
“Disjunctive normal random forests,” In Pattern Recognition, September, 2014.
DOI: 10.1016/j.patcog.2014.08.023
We develop a novel supervised learning/classification method, called disjunctive normal random forest (DNRF). A DNRF is an ensemble of randomly trained disjunctive normal decision trees (DNDT). To construct a DNDT, we formulate each decision tree in the random forest as a disjunction of rules, which are conjunctions of Boolean functions. We then approximate this disjunction of conjunctions with a differentiable function and approach the learning process as a risk minimization problem that incorporates the classification error into a single global objective function. The minimization problem is solved using gradient descent. DNRFs are able to learn complex decision boundaries and achieve low generalization error. We present experimental results demonstrating the improved performance of DNDTs and DNRFs over conventional decision trees and random forests. We also show the superior performance of DNRFs over state-of-the-art classification methods on benchmark datasets.
Keywords: Random forest, Decision tree, Classifier, Supervised learning, Disjunctive normal form


M. Seyedhosseini, T. Tasdizen.
“Scene Labeling with Contextual Hierarchical Models,” In CoRR, Vol. abs/1402.0595, 2014.
Scene labeling is the problem of assigning an object label to each pixel. It unifies the image segmentation and object recognition problems. The importance of using contextual information in scene labeling frameworks has been widely realized in the field. We propose a contextual framework, called contextual hierarchical model (CHM), which learns contextual information in a hierarchical framework for scene labeling. At each level of the hierarchy, a classifier is trained based on downsampled input images and outputs of previous levels. Our model then incorporates the resulting multi-resolution contextual information into a classifier to segment the input image at original resolution. This training strategy allows for optimization of a joint posterior probability at multiple resolutions through the hierarchy. Contextual hierarchical model is purely based on the input image patches and does not make use of any fragments or shape examples. Hence, it is applicable to a variety of problems such as object segmentation and edge detection. We demonstrate that CHM outperforms state-of-the-art on Stanford background and Weizmann horse datasets. It also outperforms state-of-the-art edge detection methods on NYU depth dataset and achieves state-of-the-art on Berkeley segmentation dataset (BSDS 500).


A. Sharma, P.T. Fletcher, J.H. Gilmore, M.L. Escolar, A. Gupta, M. Styner, G. Gerig.
“Parametric Regression Scheme for Distributions: Analysis of DTI Fiber Tract Diffusion Changes in Early Brain Development,” In Proceedings of the 2014 IEEE International Symposium on Biomedical Imaging (ISBI), pp. (accepted). 2014.
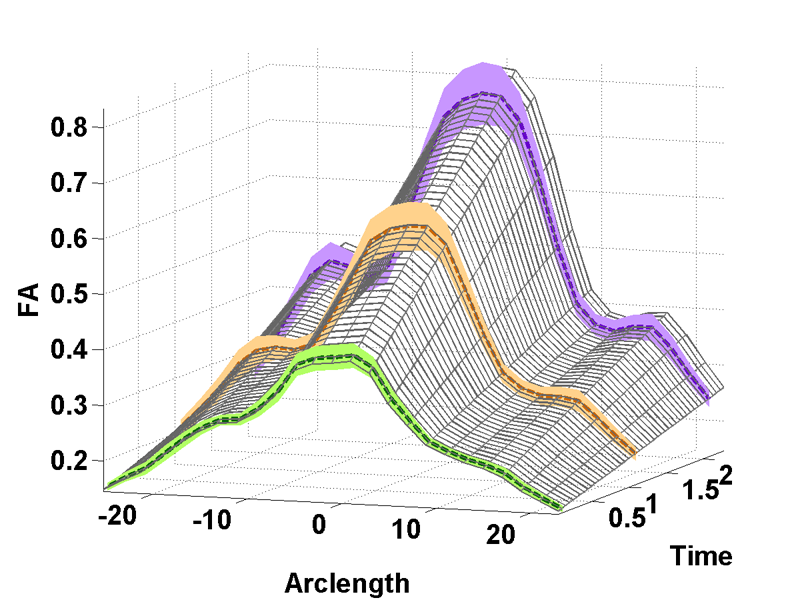
Temporal modeling frameworks often operate on scalar variables by summarizing data at initial stages as statistical summaries of the underlying distributions. For instance, DTI analysis often employs summary statistics, like mean, for regions of interest and properties along fiber tracts for population studies and hypothesis testing. This reduction via discarding of variability information may introduce significant errors which propagate through the procedures. We propose a novel framework which uses distribution-valued variables to retain and utilize the local variability information. Classic linear regression is adapted to employ these variables for model estimation. The increased stability and reliability of our proposed method when compared with regression using single-valued statistical summaries, is demonstrated in a validation experiment with synthetic data. Our driving application is the modeling of age-related changes along DTI white matter tracts. Results are shown for the spatiotemporal population trajectory of genu tract estimated from 45 healthy infants and compared with a Krabbe's patient.
Keywords: linear regression, distribution-valued data, spatiotemporal growth trajectory, DTI, early neurodevelopment


N.P. Singh, J. Hinkle, S. Joshi, P.T. Fletcher.
“An Efficient Parallel Algorithm for Hierarchical Geodesic Models in Diffeomorphisms,” In Proceedings of the 2014 IEEE International Symposium on Biomedical Imaging (ISBI), pp. (accepted). 2014.
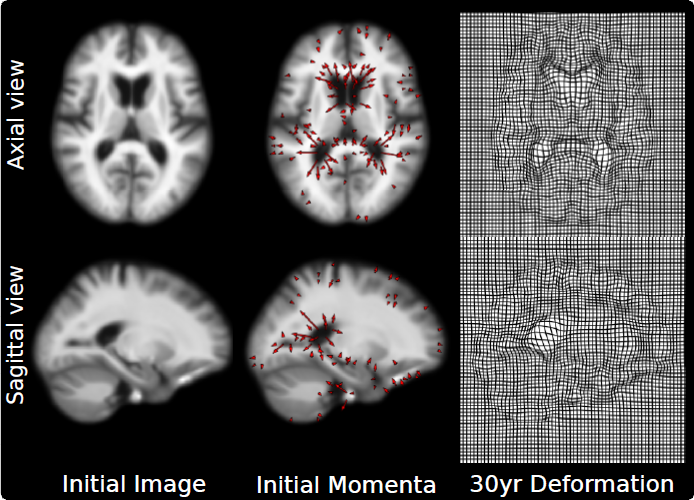
We present a novel algorithm for computing hierarchical geodesic models (HGMs) for diffeomorphic longitudinal shape analysis. The proposed algorithm exploits the inherent parallelism arising out of the independence in the contributions of individual geodesics to the group geodesic. The previous serial implementation severely limits the use of HGMs to very small population sizes due to computation time and massive memory requirements. The conventional method makes it impossible to estimate the parameters of HGMs on large datasets due to limited memory available onboard current GPU computing devices. The proposed parallel algorithm easily scales to solve HGMs on a large collection of 3D images of several individuals. We demonstrate its effectiveness on longitudinal datasets of synthetically generated shapes and 3D magnetic resonance brain images (MRI).
Keywords: LDDMM, HGM, Vector Momentum, Diffeomorphisms, Longitudinal Analysis


P. Skraba, Bei Wang, G. Chen, P. Rosen.
“2D Vector Field Simplification Based on Robustness,” In Proceedings of the 2014 IEEE Pacific Visualization Symposium, PacificVis, Note: Awarded Best Paper!, 2014.
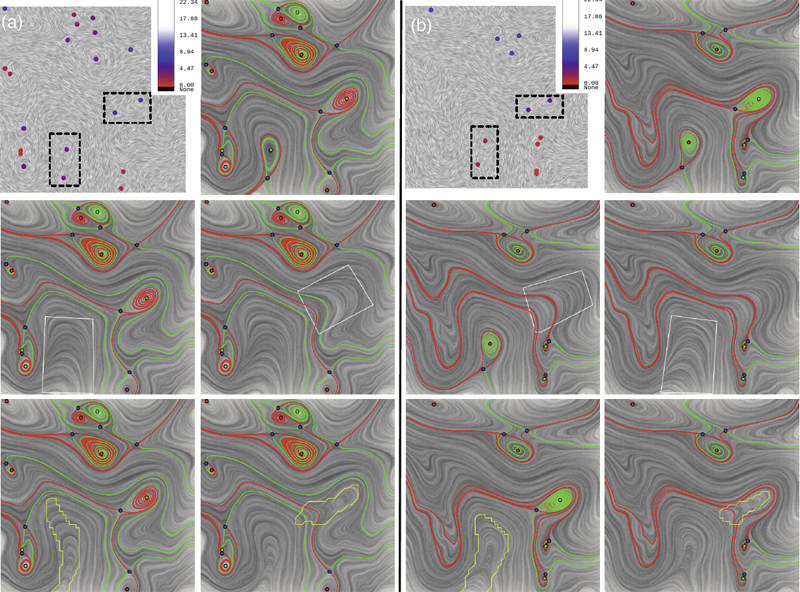
Vector field simplification aims to reduce the complexity of the flow by removing features in order of their relevance and importance, to reveal prominent behavior and obtain a compact representation for interpretation. Most existing simplification techniques based on the topological skeleton successively remove pairs of critical points connected by separatrices, using distance or area-based relevance measures. These methods rely on the stable extraction of the topological skeleton, which can be difficult due to instability in numerical integration, especially when processing highly rotational flows. These geometric metrics do not consider the flow magnitude, an important physical property of the flow. In this paper, we propose a novel simplification scheme derived from the recently introduced topological notion of robustness, which provides a complementary view on flow structure compared to the traditional topological-skeleton-based approaches. Robustness enables the pruning of sets of critical points according to a quantitative measure of their stability, that is, the minimum amount of vector field perturbation required to remove them. This leads to a hierarchical simplification scheme that encodes flow magnitude in its perturbation metric. Our novel simplification algorithm is based on degree theory, has fewer boundary restrictions, and so can handle more general cases. Finally, we provide an implementation under the piecewise-linear setting and apply it to both synthetic and real-world datasets.
Keywords: vector field, topology-based techniques, flow visualization


P. Skraba, Bei Wang.
“Interpreting Feature Tracking Through the Lens of Robustness,” In Mathematics and Visualization, Springer, pp. 19-37. 2014.
DOI: 10.1007/978-3-319-04099-8_2
A key challenge in the study of a time-varying vector fields is to resolve the correspondences between features in successive time steps and to analyze the dynamic behaviors of such features, so-called feature tracking. Commonly tracked features, such as volumes, areas, contours, boundaries, vortices, shock waves and critical points, represent interesting properties or structures of the data. Recently, the topological notion of robustness, a relative of persistent homology, has been introduced to quantify the stability of critical points. Intuitively, the robustness of a critical point is the minimum amount of perturbation necessary to cancel it. In this chapter, we offer a fresh interpretation of the notion of feature tracking, in particular, critical point tracking, through the lens of robustness.We infer correspondences between critical points based on their closeness in stability, measured by robustness, instead of just distance proximities within the domain. We prove formally that robustness helps us understand the sampling conditions under which we can resolve the correspondence problem based on region overlap techniques, and the uniqueness and uncertainty associated with such techniques. These conditions also give a theoretical basis for visualizing the piecewise linear realizations of critical point trajectories over time.


P. Skraba, Bei Wang.
“Approximating Local Homology from Samples,” In Proceedings 25th Annual ACM-SIAM Symposium on Discrete Algorithms (SODA), pp. 174-192. 2014.
Recently, multi-scale notions of local homology (a variant of persistent homology) have been used to study the local structure of spaces around a given point from a point cloud sample. Current reconstruction guarantees rely on constructing embedded complexes which become diffcult to construct in higher dimensions. We show that the persistence diagrams used for estimating local homology can be approximated using families of Vietoris-Rips complexes, whose simpler construction are robust in any dimension. To the best of our knowledge, our results, for the first time make applications based on local homology, such as stratification learning, feasible in high dimensions.

R. Stoll, E. Pardyjak, J.J. Kim, T. Harman, A.N. Hayati.
“An inter-model comparison of three computation fluid dynamics techniques for step-up and step-down street canyon flows,” In ASME FEDSM/ICNMM symposium on urban fluid mechanics, August, 2014.


M. Streit, A. Lex, S. Gratzl, C. Partl, D. Schmalstieg, H. Pfister, P. J. Park,, N. Gehlenborg.
“Guided visual exploration of genomic stratifications in cancer,” In Nature Methods, Vol. 11, No. 9, pp. 884--885. Sep, 2014.
ISSN: 1548-7091
DOI: 10.1038/nmeth.3088


B. Summa, A.A. Gooch, G. Scorzelli, V. Pascucci.
“Towards Paint and Click: Unified Interactions for Image Boundaries,” SCI Technical Report, No. UUSCI-2014-004, SCI Institute, University of Utah, December, 2014.
Image boundaries are a fundamental component of many interactive digital photography techniques, enabling applications such as segmentation, panoramas, and seamless image composition. Interactions for image boundaries often rely on two complimentary but separate approaches: editing via painting or clicking constraints. In this work, we provide a novel, unified approach for interactive editing of pairwise image boundaries that combines the ease of painting with the direct control of constraints. Rather than a sequential coupling, this new formulation allows full use of both interactions simultaneously, giving users unprecedented flexibility for fast boundary editing. To enable this new approach, we provide technical advancements. In particular, we detail a reformulation of image boundaries as a problem of finding cycles, expanding and correcting limitations of the previous work. Our new formulation provides boundary solutions for painted regions with performance on par with state-of-the-art specialized, paint-only techniques. In addition, we provide instantaneous exploration of the boundary solution space with user constraints. Furthermore, we show how to increase performance and decrease memory consumption through novel strategies and/or optional approximations. Finally, we provide examples of common graphics applications impacted by our new approach.

T. Tasdizen, M. Seyedhosseini, T. Liu, C. Jones, E. Jurrus.
“Image Segmentation for Connectomics Using Machine Learning,” In Computational Intelligence in Biomedical Imaging, Edited by Suzuki, Kenji, Springer New York, pp. 237--278. 2014.
ISBN: 978-1-4614-7244-5
DOI: 10.1007/978-1-4614-7245-2_10
Reconstruction of neural circuits at the microscopic scale of individual neurons and synapses, also known as connectomics, is an important challenge for neuroscience. While an important motivation of connectomics is providing anatomical ground truth for neural circuit models, the ability to decipher neural wiring maps at the individual cell level is also important in studies of many neurodegenerative diseases. Reconstruction of a neural circuit at the individual neuron level requires the use of electron microscopy images due to their extremely high resolution. Computational challenges include pixel-by-pixel annotation of these images into classes such as cell membrane, mitochondria and synaptic vesicles and the segmentation of individual neurons. State-of-the-art image analysis solutions are still far from the accuracy and robustness of human vision and biologists are still limited to studying small neural circuits using mostly manual analysis. In this chapter, we describe our image analysis pipeline that makes use of novel supervised machine learning techniques to tackle this problem.