SCI Publications
2013
S. Kumar, A. Saha, V. Vishwanath, P. Carns, J.A. Schmidt, G. Scorzelli, H. Kolla, R. Grout, R. Latham, R. Ross, M.E. Papka, J. Chen, V. Pascucci.
“Characterization and modeling of PIDX parallel I/O for performance optimization,” In Proceedings of SC13: International Conference for High Performance Computing, Networking, Storage and Analysis, pp. 67. 2013.
Parallel I/O library performance can vary greatly in response to user-tunable parameter values such as aggregator count, file count, and aggregation strategy. Unfortunately, manual selection of these values is time consuming and dependent on characteristics of the target machine, the underlying file system, and the dataset itself. Some characteristics, such as the amount of memory per core, can also impose hard constraints on the range of viable parameter values. In this work we address these problems by using machine learning techniques to model the performance of the PIDX parallel I/O library and select appropriate tunable parameter values. We characterize both the network and I/O phases of PIDX on a Cray XE6 as well as an IBM Blue Gene/P system. We use the results of this study to develop a machine learning model for parameter space exploration and performance prediction.
Keywords: I/O, Network Characterization, Performance Modeling
A. Lex, C. Partl, D. Kalkofen, M. Streit, A. Wasserman, S. Gratzl, D. Schmalstieg, H. Pfister.
“Entourage: Visualizing Relationships between Biological Pathways using Contextual Subsets,” In IEEE Transactions on Visualization and Computer Graphics (InfoVis '13), Vol. 19, No. 12, pp. 2536--2545. 2013.
ISSN: 1077-2626
DOI: 10.1109/TVCG.2013.154
Biological pathway maps are highly relevant tools for many tasks in molecular biology. They reduce the complexity of the overall biological network by partitioning it into smaller manageable parts. While this reduction of complexity is their biggest strength, it is, at the same time, their biggest weakness. By removing what is deemed not important for the primary function of the pathway, biologists lose the ability to follow and understand cross-talks between pathways. Considering these cross-talks is, however, critical in many analysis scenarios, such as, judging effects of drugs.
In this paper we introduce Entourage, a novel visualization technique that provides contextual information lost due to the artificial partitioning of the biological network, but at the same time limits the presented information to what is relevant to the analyst's task. We use one pathway map as the focus of an analysis and allow a larger set of contextual pathways. For these context pathways we only show the contextual subsets, i.e., the parts of the graph that are relevant to a selection. Entourage suggests related pathways based on similarities and highlights parts of a pathway that are interesting in terms of mapped experimental data. We visualize interdependencies between pathways using stubs of visual links, which we found effective yet not obtrusive. By combining this approach with visualization of experimental data, we can provide domain experts with a highly valuable tool.
We demonstrate the utility of Entourage with case studies conducted with a biochemist who researches the effects of drugs on pathways. We show that the technique is well suited to investigate interdependencies between pathways and to analyze, understand, and predict the effect that drugs have on different cell types.
T. Liu, M. Seyedhosseini, M. Ellisman, T. Tasdizen.
“Watershed Merge Forest Classification for Electron Microscopy Image Stack Segmentation,” In Proceedings of the 2013 International Conference on Image Processing, 2013.
Automated electron microscopy (EM) image analysis techniques can be tremendously helpful for connectomics research. In this paper, we extend our previous work [1] and propose a fully automatic method to utilize inter-section information for intra-section neuron segmentation of EM image stacks. A watershed merge forest is built via the watershed transform with each tree representing the region merging hierarchy of one 2D section in the stack. A section classifier is learned to identify the most likely region correspondence between adjacent sections. The inter-section information from such correspondence is incorporated to update the potentials of tree nodes. We resolve the merge forest using these potentials together with consistency constraints to acquire the final segmentation of the whole stack. We demonstrate that our method leads to notable segmentation accuracy improvement by experimenting with two types of EM image data sets.
Y. Livnat, E. Jurrus, A.V. Gundlapalli, P. Gestland.
“The CommonGround visual paradigm for biosurveillance,” In Proceedings of the 2013 IEEE International Conference on Intelligence and Security Informatics (ISI), pp. 352--357. 2013.
ISBN: 978-1-4673-6214-6
DOI: 10.1109/ISI.2013.6578857
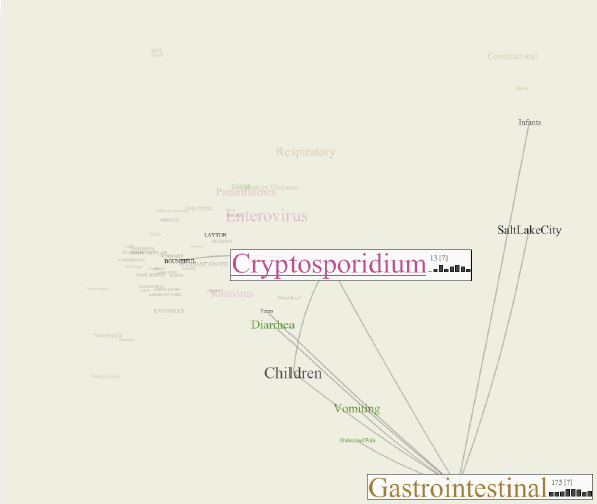
Keywords: biosurveillance, visualization, interactive exploration, situational awareness
D. Maljovec, Bei Wang, V. Pascucci, P.-T. Bremer, M.Pernice, D. Mandelli, R. Nourgaliev.
“Exploration of High-Dimensional Scalar Function for Nuclear Reactor Safety Analysis and Visualization,” In Proceedings of the 2013 International Conference on Mathematics and Computational Methods Applied to Nuclear Science & Engineering (M&C), pp. 712-723. 2013.

Keywords: high-dimensional data analysis, computational topology, nuclear reactor safety analysis, visualization
D. Maljovec, Bei Wang, D. Mandelli, P.-T. Bremer, V. Pascucci.
“Adaptive Sampling Algorithms for Probabilistic Risk Assessment of Nuclear Simulations,” In Proceedings of the 2013 International Topical Meeting on Probabilistic Safety Assessment and Analysis (PSA 2013), Note: First runner-up for Best Student Paper Award, 2013.
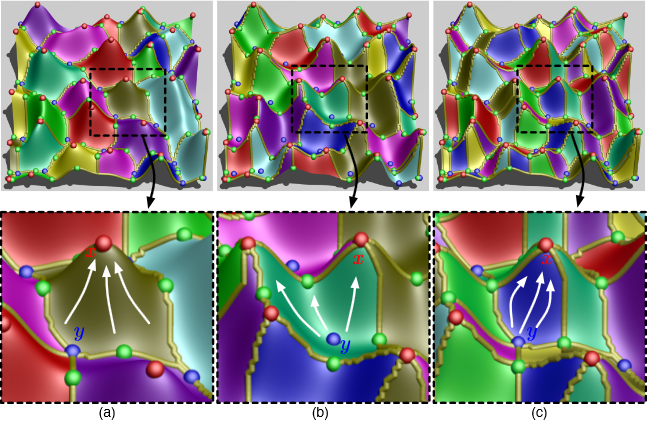
Nuclear simulations are often computationally expensive, time-consuming, and high-dimensional with respect to the number of input parameters. Thus exploring the space of all possible simulation outcomes is infeasible using finite computing resources. During simulation-based probabilistic risk analysis, it is important to discover the relationship between a potentially large number of input parameters and the output of a simulation using as few simulation trials as possible. This is a typical context for performing adaptive sampling where a few observations are obtained from the simulation, a surrogate model is built to represent the simulation space, and new samples are selected based on the model constructed. The surrogate model is then updated based on the simulation results of the sampled points. In this way, we attempt to gain the most information possible with a small number of carefully selected sampled points, limiting the number of expensive trials needed to understand features of the simulation space.
We analyze the specific use case of identifying the limit surface, i.e., the boundaries in the simulation space between system failure and system success. In this study, we explore several techniques for adaptively sampling the parameter space in order to reconstruct the limit surface. We focus on several adaptive sampling schemes. First, we seek to learn a global model of the entire simulation space using prediction models or neighborhood graphs and extract the limit surface as an iso-surface of the global model. Second, we estimate the limit surface by sampling in the neighborhood of the current estimate based on topological segmentations obtained locally.
Our techniques draw inspirations from topological structure known as the Morse-Smale complex. We highlight the advantages and disadvantages of using a global prediction model versus local topological view of the simulation space, comparing several different strategies for adaptive sampling in both contexts. One of the most interesting models we propose attempt to marry the two by obtaining a coarse global representation using prediction models, and a detailed local representation based on topology. Our methods are validated on several analytical test functions as well as a small nuclear simulation dataset modeled after a simplified Pressurized Water Reactor.
Keywords: high-dimensional data analysis, computational topology, nuclear reactor safety analysis, visualization
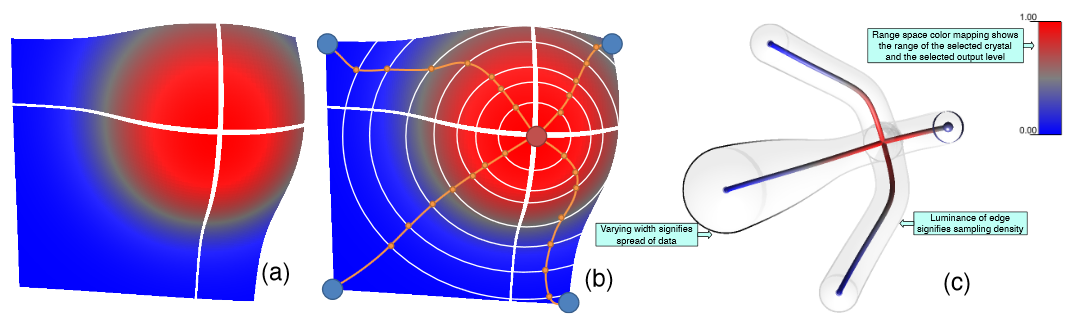
D. Maljovec, Bei Wang, D. Mandelli, P.-T. Bremer, V. Pascucci.
“Analyze Dynamic Probabilistic Risk Assessment Data through Clustering,” In Proceedings of the 2013 International Topical Meeting on Probabilistic Safety Assessment and Analysis (PSA 2013), 2013.
D. Maljovec, Bei Wang, A. Kupresanin, G. Johannesson, V. Pascucci, P.-T. Bremer.
“Adaptive Sampling with Topological Scores,” In Int. J. Uncertainty Quantification, Vol. 3, No. 2, Begell House, pp. 119--141. 2013.
DOI: 10.1615/int.j.uncertaintyquantification.2012003955
Understanding and describing expensive black box functions such as physical simulations is a common problem in many application areas. One example is the recent interest in uncertainty quantification with the goal of discovering the relationship between a potentially large number of input parameters and the output of a simulation. Typically, the simulation of interest is expensive to evaluate and thus the sampling of the parameter space is necessarily small. As a result choosing a "good" set of samples at which to evaluate is crucial to glean as much information as possible from the fewest samples. While space-filling sampling designs such as Latin hypercubes provide a good initial cover of the entire domain, more detailed studies typically rely on adaptive sampling: Given an initial set of samples, these techniques construct a surrogate model and use it to evaluate a scoring function which aims to predict the expected gain from evaluating a potential new sample. There exist a large number of different surrogate models as well as different scoring functions each with their own advantages and disadvantages. In this paper we present an extensive comparative study of adaptive sampling using four popular regression models combined with six traditional scoring functions compared against a space-filling design. Furthermore, for a single high-dimensional output function, we introduce a new class of scoring functions based on global topological rather than local geometric information. The new scoring functions are competitive in terms of the root mean squared prediction error but are expected to better recover the global topological structure. Our experiments suggest that the most common point of failure of adaptive sampling schemes are ill-suited regression models. Nevertheless, even given well-fitted surrogate models many scoring functions fail to outperform a space-filling design.
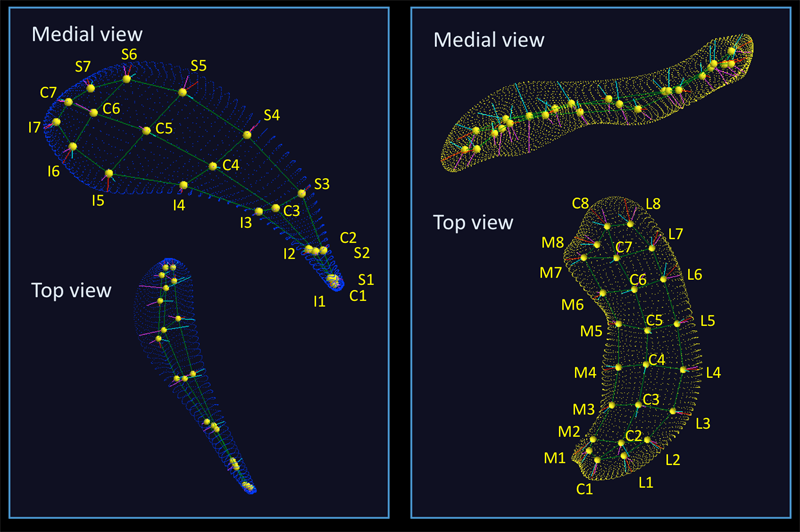
R.K. McClure, M. Styner, J.A. Lieberman, S. Gouttard, G. Gerig, X. Shi, H. Zhu.
“Localized differences in caudate and hippocampal shape associated with schizophrenia but not antipsychotic type,” In Psychiatry Research: Neuroimaging, Vol. 211, No. 1, pp. 1--10. January, 2013.
DOI: 10.1016/j.pscychresns.2012.07.001
PubMed Central ID: PMC3557605
K.S. McDowell, F. Vadakkumpadan, R. Blake, J. Blauer, G.t Plank, R.S. MacLeod, N.A. Trayanova.
“Mechanistic Inquiry into the Role of Tissue Remodeling in Fibrotic Lesions in Human Atrial Fibrillation,” In Biophysical Journal, Vol. 104, pp. 2764--2773. 2013.
DOI: 10.1016/j.bpj.2013.05.025
PubMed ID: 23790385
PubMed Central ID: PMC3686346
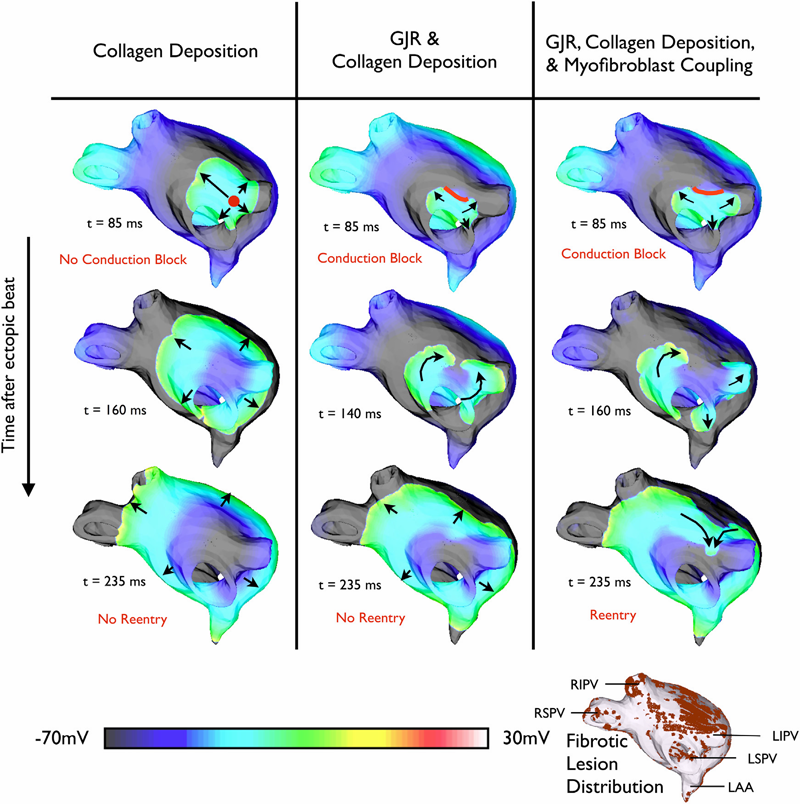
C. McGann, N. Akoum, A. Patel, E. Kholmovski, P. Revelo, K. Damal, B. Wilson, J. Cates, A. Harrison, R. Ranjan, N.S. Burgon, T. Greene, D. Kim, E.V.R. DiBella, D. Parker, R.S. MacLeod, N.F. Marrouche.
“Atrial Fibrillation Ablation Outcome is Predicted by Left Atrial Remodeling on MRI,” In Circulation: Arrhythmia and Electrophysiology, Note: Published online before print., December, 2013.
DOI: 10.1161/CIRCEP.113.000689
Background: While catheter ablation therapy for atrial fibrillation (AF) is becoming more common, results vary widely and patient selection criteria remain poorly defined. We hypothesized that late gadolinium enhancement magnetic resonance imaging (LGE-MRI) can identify left atrial (LA) wall structural remodeling (SRM) and stratify patients who are likely or not to benefit from ablation therapy.
Methods and Results: LGE-MRI was performed on 426 consecutive AF patients without contraindications to MRI and before undergoing their first ablation procedure and on 21 non-AF control subjects. Patients were categorized by SRM stage (I-IV) based on percentage of LA wall enhancement for correlation with procedure outcomes. Histological validation of SRM was performed comparing LGE-MRI to surgical biopsy. A total of 386 patients (91%) with adequate LGE-MRI scans were included in the study. Post-ablation, 123 (31.9%) experienced recurrent atrial arrhythmias over one-year follow-up. Recurrent arrhythmias (failed ablations) occurred at higher SRM stages with 28/133 (21.0%) stage I, 40/140 (29.3%) stage II, 24/71 (33.8%) stage III, and 30/42 (71.4%) stage IV. In multi-variate analysis, ablation outcome was best predicted by advanced SRM stage (hazard ratio (HR) 4.89; pKeywords: atrial fibrillation arrhythmia, catheter ablation, magnetic resonance imaging, remodeling, outcome
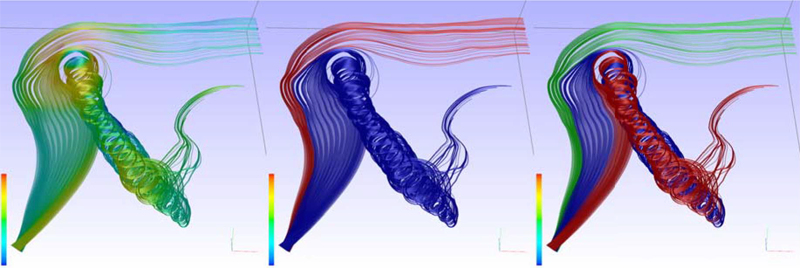
T. McLoughlin, M.W. Jones, R.S. Laramee, R. Malki, I. Masters, C.D. Hansen.
“Similarity Measures for Enhancing Interactive Streamline Seeding,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 19, No. 8, pp. 1342--1353. 2013.
ISSN: 1077-2626
DOI: 10.1109/TVCG.2012.150
PubMed ID: 23744264
Q. Meng, A. Humphrey, J. Schmidt, M. Berzins.
“Preliminary Experiences with the Uintah Framework on Intel Xeon Phi and Stampede,” SCI Technical Report, No. UUSCI-2013-002, SCI Institute, University of Utah, 2013.
In this work, we describe our preliminary experiences on the Stampede system in the context of the Uintah Computational Framework. Uintah was developed to provide an environment for solving a broad class of fluid-structure interaction problems on structured adaptive grids. Uintah uses a combination of fluid-flow solvers and particle-based methods, together with a novel asynchronous taskbased approach and fully automated load balancing. While we have designed scalable Uintah runtime systems for large CPU core counts, the emergence of heterogeneous systems presents considerable challenges in terms of effectively utilizing additional on-node accelerators and co-processors, deep memory hierarchies, as well as managing multiple levels of parallelism. Our recent work has addressed the emergence of heterogeneous CPU/GPU systems with the design of a Unified heterogeneous runtime system, enabling Uintah to fully exploit these architectures with support for asynchronous, out-of-order scheduling of both CPU and GPU computational tasks. Using this design, Uintah has run at full scale on the Keeneland System and TitanDev. With the release of the Intel Xeon Phi co-processor and the recent availability of the Stampede system, we show that Uintah may be modified to utilize such a coprocessor based system. We also explore the different usage models provided by the Xeon Phi with the aim of understanding portability of a general purpose framework like Uintah to this architecture. These usage models range from the pragma based offload model to the more complex symmetric model, utilizing all co-processor and host CPU cores simultaneously. We provide preliminary results of the various usage models for a challenging adaptive mesh refinement problem, as well as a detailed account of our experience adapting Uintah to run on the Stampede system. Our conclusion is that while the Stampede system is easy to use, obtaining high performance from the Xeon Phi co-processors requires a substantial but different investment to that needed for GPU-based systems.
Keywords: Uintah, hybrid parallelism, scalability, parallel, adaptive, MIC, Xeon Phi, heterogeneous systems, Stampede, co-processor
Q. Meng, A. Humphrey, J. Schmidt, M. Berzins.
“Investigating Applications Portability with the Uintah DAG-based Runtime System on PetaScale Supercomputers,” SCI Technical Report, No. UUSCI-2013-003, SCI Institute, University of Utah, 2013.
Present trends in high performance computing present formidable challenges for applications code using multicore nodes possibly with accelerators and/or co-processors and reduced memory while still attaining scalability. Software frameworks that execute machineindependent applications code using a runtime system that shields users from architectural complexities offer a possible solution. The Uintah framework for example, solves a broad class of large-scale problems on structured adaptive grids using fluid-flow solvers coupled with particle-based solids methods. Uintah executes directed acyclic graphs of computational tasks with a scalable asynchronous and dynamic runtime system for CPU cores and/or accelerators/coprocessors on a node. Uintah's clear separation between application and runtime code has led to scalability increases of 1000x without significant changes to application code. This methodology is tested on three leading Top500 machines; OLCF Titan, TACC Stampede and ALCF Mira using three diverse and challenging applications problems. This investigation of scalability with regard to the different processors and communications performance leads to the overall conclusion that the adaptive DAG-based approach provides a very powerful abstraction for solving challenging multiscale multi-physics engineering problems on some of the largest and most powerful computers available today.
Keywords: Uintah, hybrid parallelism, scalability, parallel, adaptive, MIC, Xeon Phi, heterogeneous systems, Stampede, co-processor
Q. Meng, A. Humphrey, J. Schmidt, M. Berzins.
“Investigating Applications Portability with the Uintah DAG-based Runtime System on PetaScale Supercomputers,” In Proceedings of SC13: International Conference for High Performance Computing, Networking, Storage and Analysis, pp. 96:1--96:12. 2013.
ISBN: 978-1-4503-2378-9
DOI: 10.1145/2503210.2503250
Present trends in high performance computing present formidable challenges for applications code using multicore nodes possibly with accelerators and/or co-processors and reduced memory while still attaining scalability. Software frameworks that execute machine-independent applications code using a runtime system that shields users from architectural complexities offer a possible solution. The Uintah framework for example, solves a broad class of large-scale problems on structured adaptive grids using fluid-flow solvers coupled with particle-based solids methods. Uintah executes directed acyclic graphs of computational tasks with a scalable asynchronous and dynamic runtime system for CPU cores and/or accelerators/co-processors on a node. Uintah's clear separation between application and runtime code has led to scalability increases of 1000x without significant changes to application code. This methodology is tested on three leading Top500 machines; OLCF Titan, TACC Stampede and ALCF Mira using three diverse and challenging applications problems. This investigation of scalability with regard to the different processors and communications performance leads to the overall conclusion that the adaptive DAG-based approach provides a very powerful abstraction for solving challenging multi-scale multi-physics engineering problems on some of the largest and most powerful computers available today.
Keywords: Blue Gene/Q, GPU, Xeon Phi, adaptive, application, co-processor, heterogeneous systems, hybrid parallelism, parallel, scalability, software, uintah, NETL
Q. Meng, A. Humphrey, J. Schmidt, M. Berzins.
“Preliminary Experiences with the Uintah Framework on Intel Xeon Phi and Stampede,” In Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery (XSEDE 2013), San Diego, California, pp. 48:1--48:8. 2013.
DOI: 10.1145/2484762.2484779
In this work, we describe our preliminary experiences on the Stampede system in the context of the Uintah Computational Framework. Uintah was developed to provide an environment for solving a broad class of fluid-structure interaction problems on structured adaptive grids. Uintah uses a combination of fluid-flow solvers and particle-based methods, together with a novel asynchronous task-based approach and fully automated load balancing. While we have designed scalable Uintah runtime systems for large CPU core counts, the emergence of heterogeneous systems presents considerable challenges in terms of effectively utilizing additional on-node accelerators and co-processors, deep memory hierarchies, as well as managing multiple levels of parallelism. Our recent work has addressed the emergence of heterogeneous CPU/GPU systems with the design of a Unified heterogeneous runtime system, enabling Uintah to fully exploit these architectures with support for asynchronous, out-of-order scheduling of both CPU and GPU computational tasks. Using this design, Uintah has run at full scale on the Keeneland System and TitanDev. With the release of the Intel Xeon Phi co-processor and the recent availability of the Stampede system, we show that Uintah may be modified to utilize such a co-processor based system. We also explore the different usage models provided by the Xeon Phi with the aim of understanding portability of a general purpose framework like Uintah to this architecture. These usage models range from the pragma based offload model to the more complex symmetric model, utilizing all co-processor and host CPU cores simultaneously. We provide preliminary results of the various usage models for a challenging adaptive mesh refinement problem, as well as a detailed account of our experience adapting Uintah to run on the Stampede system. Our conclusion is that while the Stampede system is easy to use, obtaining high performance from the Xeon Phi co-processors requires a substantial but different investment to that needed for GPU-based systems.
Keywords: MIC, Xeon Phi, adaptive, co-processor, heterogeneous systems, hybrid parallelism, parallel, scalability, stampede, uintah, c-safe
D.C.B. de Oliveira, Z. Rakamaric, G. Gopalakrishnan, A. Humphrey, Q. Meng, M. Berzins.
“Crash Early, Crash Often, Explain Well: Practical Formal Correctness Checking of Million-core Problem Solving Environments for HPC,” In Proceedings of the 35th International Conference on Software Engineering (ICSE 2013), pp. (accepted). 2013.
While formal correctness checking methods have been deployed at scale in a number of important practical domains, we believe that such an experiment has yet to occur in the domain of high performance computing at the scale of a million CPU cores. This paper presents preliminary results from the Uintah Runtime Verification (URV) project that has been launched with this objective. Uintah is an asynchronous task-graph based problem-solving environment that has shown promising results on problems as diverse as fluid-structure interaction and turbulent combustion at well over 200K cores to date. Uintah has been tested on leading platforms such as Kraken, Keenland, and Titan consisting of multicore CPUs and GPUs, incorporates several innovative design features, and is following a roadmap for development well into the million core regime. The main results from the URV project to date are crystallized in two observations: (1) A diverse array of well-known ideas from lightweight formal methods and testing/observing HPC systems at scale have an excellent chance of succeeding. The real challenges are in finding out exactly which combinations of ideas to deploy, and where. (2) Large-scale problem solving environments for HPC must be designed such that they can be \"crashed early\" (at smaller scales of deployment) and \"crashed often\" (have effective ways of input generation and schedule perturbation that cause vulnerabilities to be attacked with higher probability). Furthermore, following each crash, one must \"explain well\" (given the extremely obscure ways in which an error finally manifests itself, we must develop ways to record information leading up to the crash in informative ways, to minimize offsite debugging burden). Our plans to achieve these goals and to measure our success are described. We also highlight some of the broadly applicable concepts and approaches.
Keywords: Uintah
B. Paniagua, O. Emodi, J. Hill, J. Fishbaugh, L.A. Pimenta, S.R. Aylward, E. Andinet, G. Gerig, J. Gilmore, J.A. van Aalst, M. Styner.
“3D of brain shape and volume after cranial vault remodeling surgery for craniosynostosis correction in infants,” In Proceedings of SPIE 8672, Medical Imaging 2013: Biomedical Applications in Molecular, Structural, and Functional Imaging, 86720V, 2013.
DOI: 10.1117/12.2006524

B. Paniagua, A. Lyall, J.-B. Berger, C. Vachet, R.M. Hamer, S. Woolson, W. Lin, J. Gilmore, M. Styner.
“Lateral ventricle morphology analysis via mean latitude axis,” In Proceedings of SPIE 8672, Biomedical Applications in Molecular, Structural, and Functional Imaging, 86720M, 2013.
DOI: 10.1117/12.2006846
PubMed ID: 23606800
PubMed Central ID: PMC3630372
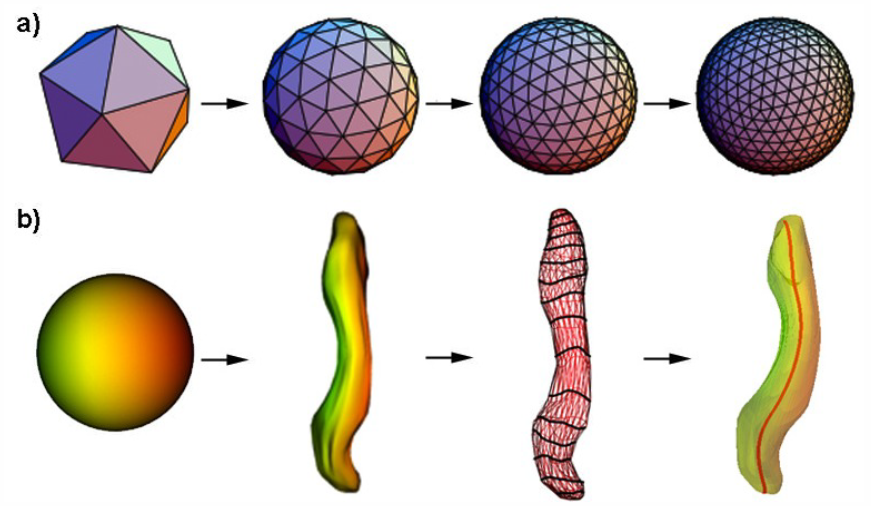
C. Partl, A. Lex, M. Streit, D. Kalkofen, K. Kashofer, D. Schmalstieg.
“enRoute: Dynamic Path Extraction from Biological Pathway Maps for Exploring Heterogeneous Experimental Datasets,” In BMC Bioinformatics, Vol. 14, No. Suppl 19, Nov, 2013.
ISSN: 1471-2105
DOI: 10.1186/1471-2105-14-S19-S3
Jointly analyzing biological pathway maps and experimental data is critical for understanding how biological processes work in different conditions and why different samples exhibit certain characteristics. This joint analysis, however, poses a significant challenge for visualization. Current techniques are either well suited to visualize large amounts of pathway node attributes, or to represent the topology of the pathway well, but do not accomplish both at the same time. To address this we introduce enRoute, a technique that enables analysts to specify a path of interest in a pathway, extract this path into a separate, linked view, and show detailed experimental data associated with the nodes of this extracted path right next to it. This juxtaposition of the extracted path and the experimental data allows analysts to simultaneously investigate large amounts of potentially heterogeneous data, thereby solving the problem of joint analysis of topology and node attributes. As this approach does not modify the layout of pathway maps, it is compatible with arbitrary graph layouts, including those of hand-crafted, image-based pathway maps. We demonstrate the technique in context of pathways from the KEGG and the Wikipathways databases. We apply experimental data from two public databases, the Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) that both contain a wide variety of genomic datasets for a large number of samples. In addition, we make use of a smaller dataset of hepatocellular carcinoma and common xenograft models. To verify the utility of enRoute, domain experts conducted two case studies where they explore data from the CCLE and the hepatocellular carcinoma datasets in the context of relevant pathways.
Page 47 of 144
