SCI Publications
2015


P. Skraba, Bei Wang, G. Chen, P. Rosen.
“Robustness-Based Simplification of 2D Steady and Unsteady Vector Fields,” In IEEE Transactions on Visualization and Computer Graphics (to appear), 2015.
Vector field simplification aims to reduce the complexity of the flow by removing features in order of their relevance and importance, to reveal prominent behavior and obtain a compact representation for interpretation. Most existing simplification techniques based on the topological skeleton successively remove pairs of critical points connected by separatrices, using distance or area-based relevance measures. These methods rely on the stable extraction of the topological skeleton, which can be difficult due to instability in numerical integration, especially when processing highly rotational flows. In this paper, we propose a novel simplification scheme derived from the recently introduced topological notion of robustness which enables the pruning of sets of critical points according to a quantitative measure of their stability, that is, the minimum amount of vector field perturbation required to remove them. This leads to a hierarchical simplification scheme that encodes flow magnitude in its perturbation metric. Our novel simplification algorithm is based on degree theory and has minimal boundary restrictions. Finally, we provide an implementation under the piecewise-linear setting and apply it to both synthetic and real-world datasets. We show local and complete hierarchical simplifications for steady as well as unsteady vector fields.

SLASH.
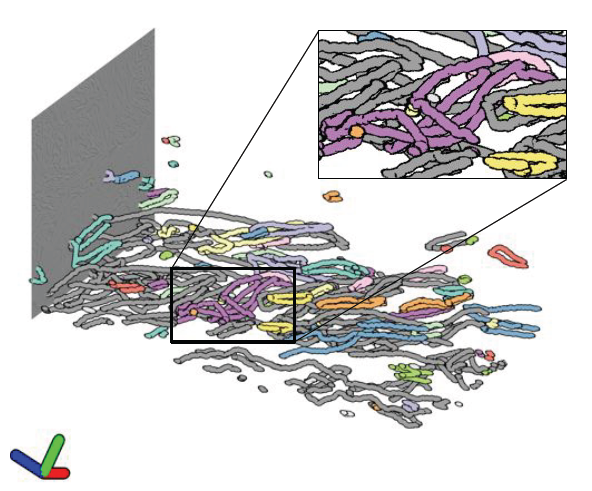
Note: SLASH: A hybrid system for high-throughput segmentation of large neuropil datasets, SLASH is funded by the National Institute of Neurological Disorders and Stroke (NINDS) grant 5R01NS075314-03., 2015.


H. Strobelt, B. Alsallakh, J. Botros, B. Peterson, M. Borowsky, H. Pfister,, A. Lex.
“Vials: Visualizing Alternative Splicing of Genes,” In IEEE Transactions on Visualization and Computer Graphics (InfoVis '15), Vol. 22, No. 1, pp. 399-408. 2015.
Alternative splicing is a process by which the same DNA sequence is used to assemble different proteins, called protein isoforms. Alternative splicing works by selectively omitting some of the coding regions (exons) typically associated with a gene. Detection of alternative splicing is difficult and uses a combination of advanced data acquisition methods and statistical inference. Knowledge about the abundance of isoforms is important for understanding both normal processes and diseases and to eventually improve treatment through targeted therapies. The data, however, is complex and current visualizations for isoforms are neither perceptually efficient nor scalable. To remedy this, we developed Vials, a novel visual analysis tool that enables analysts to explore the various datasets that scientists use to make judgments about isoforms: the abundance of reads associated with the coding regions of the gene, evidence for junctions, i.e., edges connecting the coding regions, and predictions of isoform frequencies. Vials is scalable as it allows for the simultaneous analysis of many samples in multiple groups. Our tool thus enables experts to (a) identify patterns of isoform abundance in groups of samples and (b) evaluate the quality of the data. We demonstrate the value of our tool in case studies using publicly available datasets.


B. Summa, A. A. Gooch, G. Scorzelli, V. Pascucci.
“Paint and Click: Unified Interactions for Image Boundaries,” In Computer Graphics Forum, Vol. 34, No. 2, Wiley-Blackwell, pp. 385--393. May, 2015.
DOI: 10.1111/cgf.12568
Image boundaries are a fundamental component of many interactive digital photography techniques, enabling applications such as segmentation, panoramas, and seamless image composition. Interactions for image boundaries often rely on two complementary but separate approaches: editing via painting or clicking constraints. In this work, we provide a novel, unified approach for interactive editing of pairwise image boundaries that combines the ease of painting with the direct control of constraints. Rather than a sequential coupling, this new formulation allows full use of both interactions simultaneously, giving users unprecedented flexibility for fast boundary editing. To enable this new approach, we provide technical advancements. In particular, we detail a reformulation of image boundaries as a problem of finding cycles, expanding and correcting limitations of the previous work. Our new formulation provides boundary solutions for painted regions with performance on par with state-of-the-art specialized, paint-only techniques. In addition, we provide instantaneous exploration of the boundary solution space with user constraints. Finally, we provide examples of common graphics applications impacted by our new approach.


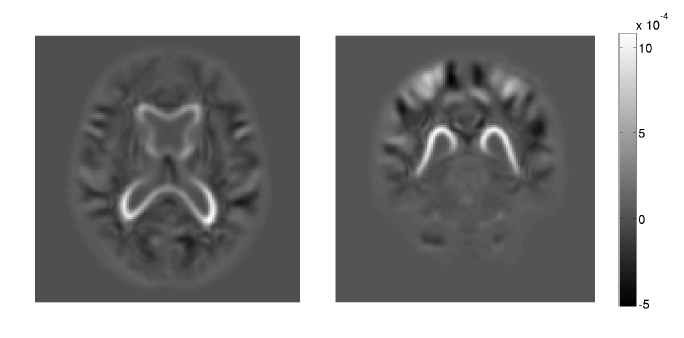
M. R. Swanson, J. J. Wolff, J. T. Elison, H. Gu, H. C. Hazlett, K. Botteron, M. Styner, S. Paterson, G. Gerig, J. Constantino, S. Dager, A. Estes, C. Vachet, J. Piven.
“Splenium development and early spoken language in human infants,” In Developmental Science, Wiley Online Library, 2015.
ISSN: 1467-7687
DOI: 10.1111/desc.12360
The association between developmental trajectories of language-related white matter fiber pathways from 6 to 24 months of age and individual differences in language production at 24 months of age was investigated. The splenium of the corpus callosum, a fiber pathway projecting through the posterior hub of the default mode network to occipital visual areas, was examined as well as pathways implicated in language function in the mature brain, including the arcuate fasciculi, uncinate fasciculi, and inferior longitudinal fasciculi. The hypothesis that the development of neural circuitry supporting domain-general orienting skills would relate to later language performance was tested in a large sample of typically developing infants. The present study included 77 infants with diffusion weighted MRI scans at 6, 12 and 24 months and language assessment at 24 months. The rate of change in splenium development varied significantly as a function of language production, such that children with greater change in fractional anisotropy (FA) from 6 to 24 months produced more words at 24 months. Contrary to findings from older children and adults, significant associations between language production and FA in the arcuate, uncinate, or left inferior longitudinal fasciculi were not observed. The current study highlights the importance of tracing brain development trajectories from infancy to fully elucidate emerging brain–behavior associations while also emphasizing the role of the splenium as a key node in the structural network that supports the acquisition of spoken language.

Note: VisTrails: A scientific workflow management system. Scientific Computing and Imaging Institute (SCI), Download from: http://www.vistrails.org, 2015.


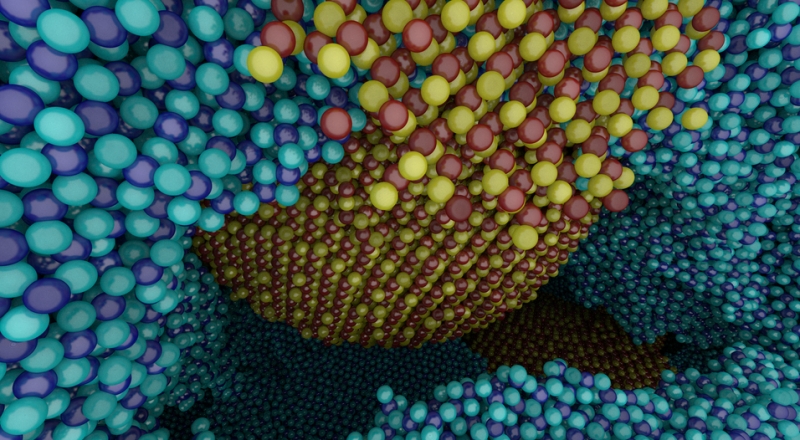
I. Wald, A. Knoll, G. P. Johnson, W. Usher, V. Pascucci, M. E. Papka.
“CPU Ray Tracing Large Particle Data with Balanced P-k-d Trees,” In 2015 IEEE Scientific Visualization Conference, IEEE, Oct, 2015.
DOI: 10.1109/scivis.2015.7429492
We present a novel approach to rendering large particle data sets from molecular dynamics, astrophysics and other sources. We employ a new data structure adapted from the original balanced k-d tree, which allows for representation of data with trivial or no overhead. In the OSPRay visualization framework, we have developed an efficient CPU algorithm for traversing, classifying and ray tracing these data. Our approach is able to render up to billions of particles on a typical workstation, purely on the CPU, without any approximations or level-of-detail techniques, and optionally with attribute-based color mapping, dynamic range query, and advanced lighting models such as ambient occlusion and path tracing.


R. Whitaker, W. Thompson, J. Berger, B. Fischhof, M. Goodchild, M. Hegarty, C. Jermaine, K. S. McKinley, A. Pang, J. Wendelberger.
“Workshop on Quantification, Communication, and Interpretation of Uncertainty in Simulation and Data Science,” Note: Computing Community Consortium, 2015.
Modern science, technology, and politics are all permeated by data that comes from people, measurements, or computational processes. While this data is often incomplete, corrupt, or lacking in sufficient accuracy and precision, explicit consideration of uncertainty is rarely part of the computational and decision making pipeline. The CCC Workshop on Quantification, Communication, and Interpretation of Uncertainty in Simulation and Data Science explored this problem, identifying significant shortcomings in the ways we currently process, present, and interpret uncertain data. Specific recommendations on a research agenda for the future were made in four areas: uncertainty quantification in large-scale computational simulations, uncertainty quantification in data science, software support for uncertainty computation, and better integration of uncertainty quantification and communication to stakeholders.


J. J. Wolff, G. Gerig, J. D. Lewis, T. Soda, M. A. Styner, C. Vachet, K. N. Botteron, J. T. Elison, S. R. Dager, A. M. Estes, H. C. Hazlett, R. T. Schultz, L. Zwaigenbaum, J. Piven.
“Altered corpus callosum morphology associated with autism over the first 2 years of life,” In Brain, 2015.
DOI: 10.1093/brain/awv118
Numerous brain imaging studies indicate that the corpus callosum is smaller in older children and adults with autism spectrum disorder. However, there are no published studies examining the morphological development of this connective pathway in infants at-risk for the disorder. Magnetic resonance imaging data were collected from 270 infants at high familial risk for autism spectrum disorder and 108 low-risk controls at 6, 12 and 24 months of age, with 83% of infants contributing two or more data points. Fifty-seven children met criteria for ASD based on clinical-best estimate diagnosis at age 2 years. Corpora callosa were measured for area, length and thickness by automated segmentation. We found significantly increased corpus callosum area and thickness in children with autism spectrum disorder starting at 6 months of age. These differences were particularly robust in the anterior corpus callosum at the 6 and 12 month time points. Regression analysis indicated that radial diffusivity in this region, measured by diffusion tensor imaging, inversely predicted thickness. Measures of area and thickness in the first year of life were correlated with repetitive behaviours at age 2 years. In contrast to work from older children and adults, our findings suggest that the corpus callosum may be larger in infants who go on to develop autism spectrum disorder. This result was apparent with or without adjustment for total brain volume. Although we did not see a significant interaction between group and age, cross-sectional data indicated that area and thickness differences diminish by age 2 years. Regression data incorporating diffusion tensor imaging suggest that microstructural properties of callosal white matter, which includes myelination and axon composition, may explain group differences in morphology.


M. Zhang, P. T. Fletcher.
“Finite-Dimensional Lie Algebras for Fast Diffeomorphic Image Registration,” In Information Processing in Medical Imaging (IPMI), 2015.
This paper presents a fast geodesic shooting algorithm for diffeomorphic image registration. We first introduce a novel finite-dimensional Lie algebra structure on the space of bandlimited velocity fields. We then show that this space can effectively represent initial velocities for diffeomorphic image registration at much lower dimensions than typically used, with little to no loss in registration accuracy. We then leverage the fact that the geodesic evolution equations, as well as the adjoint Jacobi field equations needed for gradient descent methods, can be computed entirely in this finite-dimensional Lie algebra. The result is a geodesic shooting method for large deformation metric mapping (LDDMM) that is dramatically faster and less memory intensive than state-of-the-art methods. We demonstrate the effectiveness of our model to register 3D brain images and compare its registration accuracy, runtime, and memory consumption with leading LDDMM methods. We also show how our algorithm breaks through the prohibitive time and memory requirements of diffeomorphic atlas building.


M. Zhang, P. T. Fletcher.
“Bayesian Principal Geodesic Analysis for Estimating Intrinsic Diffeomorphic Image Variability,” In Medical Image Analysis (accepted), 2015.
In this paper, we present a generative Bayesian approach for estimating the low-dimensional latent space of diffeomorphic shape variability in a population of images. We develop a latent variable model for principal geodesic analysis (PGA) that provides a probabilistic framework for factor analysis in the space of diffeomorphisms. A sparsity prior in the model results in automatic selection of the number of relevant dimensions by driving unnecessary principal geodesics to zero. To infer model parameters, including the image atlas, principal geodesic deformations, and the effective dimensionality, we introduce an expectation maximization (EM) algorithm. We evaluate our proposed model on 2D synthetic data and the 3D OASIS brain database of magnetic resonance images, and show that the automatically selected latent dimensions from our model are able to reconstruct unobserved testing images with lower error than both linear principal component analysis (LPCA) in the image space and tangent space principal component analysis (TPCA) in the diffeomorphism space.


M. Zhang, H. Shao, P. T. Fletcher.
“A Mixture Model for Automatic Diffeomorphic Multi-Atlas Building,” In MICCAI Workshop, Springer, 2015.
Computing image atlases that are representative of a dataset
is an important first step for statistical analysis of images. Most current approaches estimate a single atlas to represent the average of a large population of images, however, a single atlas is not sufficiently expressive to capture distributions of images with multiple modes. In this paper, we present a mixture model for building diffeomorphic multi-atlases that can represent sub-populations without knowing the category of each observed data point. In our probabilistic model, we treat diffeomorphic image transformations as latent variables, and integrate them out using a Monte Carlo Expectation Maximization (MCEM) algorithm via Hamiltonian Monte Carlo (HMC) sampling. A key benefit of our model is that the mixture modeling inference procedure results in an automatic clustering of the dataset. Using 2D synthetic data generated from known parameters, we demonstrate the ability of our model to successfully recover the multi-atlas and automatically cluster the dataset. We also show the effectiveness of the proposed method in a multi-atlas estimation problem for 3D brain images.
2014


G. Adluru, Y. Gur, J. Anderson, L. Richards, N. Adluru, E. DiBella.
“Assessment of white matter microstructure in stroke patients using NODDI,” In Proceedings of the 2014 IEEE Int. Conf. Engineering and Biology Society (EMBC), 2014.
Diffusion weighted imaging (DWI) is widely used to study changes in white matter following stroke. In various studies employing diffusion tensor imaging (DTI) and high angular resolution diffusion imaging (HARDI) modalities, it has been shown that fractional anisotropy (FA), mean diffusivity (MD), and generalized FA (GFA) can be used as measures of white matter tract integrity in stroke patients. However, these measures may be non-specific, as they do not directly delineate changes in tissue microstructure. Multi-compartment models overcome this limitation by modeling DWI data using a set of indices that are directly related to white matter microstructure. One of these models which is gaining popularity, is neurite orientation dispersion and density imaging (NODDI). This model uses conventional single or multi-shell HARDI data to describe fiber orientation dispersion as well as densities of different tissue types in the imaging voxel. In this paper, we apply for the first time the NODDI model to 4-shell HARDI stroke data. By computing NODDI indices over the entire brain in two stroke patients, and comparing tissue regions in ipsilesional and contralesional hemispheres, we demonstrate that NODDI modeling provides specific information on tissue microstructural changes. We also introduce an information theoretic analysis framework to investigate the non-local effects of stroke in the white matter. Our initial results suggest that the NODDI indices might be more specific markers of white matter reorganization following stroke than other measures previously used in studies of stroke recovery.

S.P. Awate, R.T. Whitaker.
“Multiatlas Segmentation as Nonparametric Regression,” In IEEE Trans Med Imaging, April, 2014.
PubMed ID: 24802528
This paper proposes a novel theoretical framework to model and analyze the statistical characteristics of a wide range of segmentation methods that incorporate a database of label maps or atlases; such methods are termed as label fusion or multiatlas segmentation. We model these multiatlas segmentation problems as nonparametric regression problems in the high-dimensional space of image patches. We analyze the nonparametric estimator's convergence behavior that characterizes expected segmentation error as a function of the size of the multiatlas database. We show that this error has an analytic form involving several parameters that are fundamental to the specific segmentation problem (determined by the chosen anatomical structure, imaging modality, registration algorithm, and labelfusion algorithm). We describe how to estimate these parameters and show that several human anatomical structures exhibit the trends modeled analytically. We use these parameter estimates to optimize the regression estimator. We show that the expected error for large database sizes is well predicted by models learned on small databases. Thus, a few expert segmentations can help predict the database sizes required to keep the expected error below a specified tolerance level. Such cost-benefit analysis is crucial for deploying clinical multiatlas segmentation systems.

S.P. Awate, Y.-Y. Yu, R.T. Whitaker.
“Kernel Principal Geodesic Analysis,” In Proceedings of the European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML PKDD), Springer LNAI, 2014.
Kernel principal component analysis (kPCA) has been proposed as a dimensionality-reduction technique that achieves nonlinear, low-dimensional representations of data via the mapping to kernel feature space. Conventionally, kPCA relies on Euclidean statistics in kernel feature space. However, Euclidean analysis can make kPCA inefficient or incorrect for many popular kernels that map input points to a hypersphere in kernel feature space. To address this problem, this paper proposes a novel adaptation of kPCA, namely kernel principal geodesic analysis (kPGA), for hyperspherical statistical analysis in kernel feature space. This paper proposes tools for statistical analyses on the Riemannian manifold of the Hilbert sphere in the reproducing kernel Hilbert space, including algorithms for computing the sample weighted Karcher mean and eigen analysis of the sample weighted Karcher covariance. It then applies these tools to propose novel methods for (i)~dimensionality reduction and (ii)~clustering using mixture-model fitting. The results, on simulated and real-world data, show that kPGA-based methods perform favorably relative to their kPCA-based analogs.


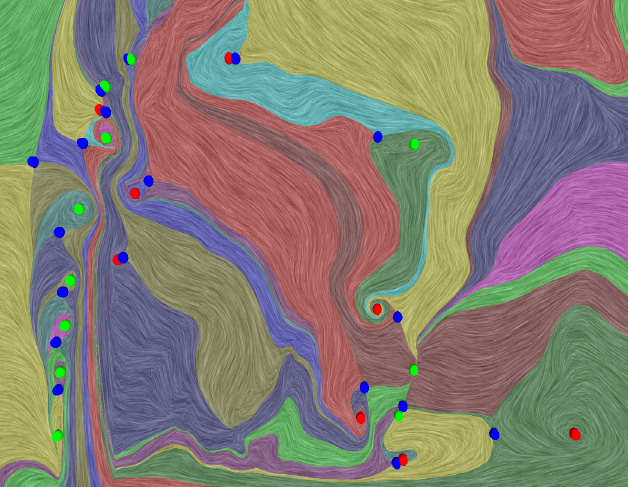
H. Bhatia, V. Pascucci, R.M. Kirby, P.-T. Bremer.
“Extracting Features from Time-Dependent Vector Fields Using Internal Reference Frames,” In Computer Graphics Forum, Vol. 33, No. 3, pp. 21--30. June, 2014.
DOI: 10.1111/cgf.12358
Extracting features from complex, time-dependent flow fields remains a significant challenge despite substantial research efforts, especially because most flow features of interest are defined with respect to a given reference frame. Pathline-based techniques, such as the FTLE field, are complex to implement and resource intensive, whereas scalar transforms, such as λ2, often produce artifacts and require somewhat arbitrary thresholds. Both approaches aim to analyze the flow in a more suitable frame, yet neither technique explicitly constructs one.
This paper introduces a new data-driven technique to compute internal reference frames for large-scale complex flows. More general than uniformly moving frames, these frames can transform unsteady fields, which otherwise require substantial processing of resources, into a sequence of individual snapshots that can be analyzed using the large body of steady-flow analysis techniques. Our approach is simple, theoretically well-founded, and uses an embarrassingly parallel algorithm for structured as well as unstructured data. Using several case studies from fluid flow and turbulent combustion, we demonstrate that internal frames are distinguished, result in temporally coherent structures, and can extract well-known as well as notoriously elusive features one snapshot at a time.


H. Bhatia, A. Gyulassy, H. Wang, P.-T. Bremer, V. Pascucci .
“Robust Detection of Singularities in Vector Fields,” In Topological Methods in Data Analysis and Visualization III, Mathematics and Visualization, Springer International Publishing, pp. 3--18. March, 2014.
DOI: 10.1007/978-3-319-04099-8_1
Recent advances in computational science enable the creation of massive datasets of ever increasing resolution and complexity. Dealing effectively with such data requires new analysis techniques that are provably robust and that generate reproducible results on any machine. In this context, combinatorial methods become particularly attractive, as they are not sensitive to numerical instabilities or the details of a particular implementation. We introduce a robust method for detecting singularities in vector fields. We establish, in combinatorial terms, necessary and sufficient conditions for the existence of a critical point in a cell of a simplicial mesh for a large class of interpolation functions. These conditions are entirely local and lead to a provably consistent and practical algorithm to identify cells containing singularities.


H. Bhatia, V. Pascucci, P.-T. Bremer.
“The Natural Helmholtz-Hodge Decomposition For Open-Boundary Flow Analysis,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 99, pp. 1566--1578. 2014.
DOI: 10.1109/TVCG.2014.2312012
The Helmholtz-Hodge decomposition (HHD) describes a flow as the sum of an incompressible, an irrotational, and a harmonic flow, and is a fundamental tool for simulation and analysis. Unfortunately, for bounded domains, the HHD is not uniquely defined, and traditionally, boundary conditions are imposed to obtain a unique solution. However, in general, the boundary conditions used during the simulation may not be known and many simulations use open boundary conditions. In these cases, the flow imposed by traditional boundary conditions may not be compatible with the given data, which leads to sometimes drastic artifacts and distortions in all three components, hence producing unphysical results. Instead, this paper proposes the natural HHD, which is defined by separating the flow into internal and external components. Using a completely data-driven approach, the proposed technique obtains uniqueness without assuming boundary conditions a priori. As a result, it enables a reliable and artifact-free analysis for flows with open boundaries or unknown boundary conditions. Furthermore, our approach computes the HHD on a point-wise basis in contrast to the existing global techniques, and thus supports computing inexpensive local approximations for any subset of the domain. Finally, the technique is easy to implement for a variety of spatial discretizations and interpolated fields in both two and three dimensions.


A. Bigelow, S. Drucker, D. Fisher, M.D. Meyer.
“Reflections on How Designers Design With Data,” In Proceedings of the ACM International Conference on Advanced Visual Interfaces (AVI), Note: Awarded Best Paper!, 2014.
In recent years many popular data visualizations have emerged that are created largely by designers whose main area of expertise is not computer science. Designers generate these visualizations using a handful of design tools and environments. To better inform the development of tools intended for designers working with data, we set out to understand designers' challenges and perspectives. We interviewed professional designers, conducted observations of designers working with data in the lab, and observed designers working with data in team settings in the wild. A set of patterns emerged from these observations from which we extract a number of themes that provide a new perspective on design considerations for visualization tool creators, as well as on known engineering problems.
Keywords: Visualization, infographics, design practice



J.J.E. Blauer, D. Swenson, K. Higuchi, G. Plank, R. Ranjan, N. Marrouche,, R.S. MacLeod.
“Sensitivity and Specificity of Substrate Mapping: An In Silico Framework for the Evaluation of Electroanatomical Substrate Mapping Strategies,” In Journal of Cardiovascular Electrophysiology, In Journal of Cardiovascular Electrophysiology, Vol. 25, No. 7, Note: Featured on journal cover., pp. 774--780. May, 2014.
Background - Voltage mapping is an important tool for characterizing proarrhythmic electrophysiological substrate, yet it is subject to geometric factors that influence bipolar amplitudes and thus compromise performance. The aim of this study was to characterize the impact of catheter orientation on the ability of bipolar amplitudes to accurately discriminate between healthy and diseased tissues.
Methods and Results - We constructed a three-dimensional, in-silico, bidomain model of cardiac tissue containing transmural lesions of varying diameter. A planar excitation wave was stimulated and electrograms were sampled with a realistic catheter model at multiple positions and orientations. We carried out validation studies in animal experiments of acute ablation lesions mapped with a clinical mapping system. Bipolar electrograms sampled at higher inclination angles of the catheter with respect to the tissue demonstrated improvements in both sensitivity and specificity of lesion detection. Removing low voltage electrograms with concurrent activation of both electrodes, suggesting false attenuation of the bipolar electrogram due to alignment with the excitation wavefront, had little effect on the accuracy of voltage mapping.
Conclusions - Our results demonstrate possible mechanisms for the impact of catheter orientation on voltage mapping accuracy. Moreover, results from our simulations suggest that mapping accuracy may be improved by selectively controlling the inclination of the catheter to record at higher angles with respect to the tissue.
Keywords: arrhythmia, computer-based model, electroanatomical mapping, voltage mapping, bipolar electrogram