Visualization
Visualization, sometimes referred to as visual data analysis, uses the graphical representation of data as a means of gaining understanding and insight into the data. Visualization research at SCI has focused on applications spanning computational fluid dynamics, medical imaging and analysis, biomedical data analysis, healthcare data analysis, weather data analysis, poetry, network and graph analysis, financial data analysis, etc.Research involves novel algorithm and technique development to building tools and systems that assist in the comprehension of massive amounts of (scientific) data. We also research the process of creating successful visualizations.
We strongly believe in the role of interactivity in visual data analysis. Therefore, much of our research is concerned with creating visualizations that are intuitive to interact with and also render at interactive rates.
Visualization at SCI includes the academic subfields of Scientific Visualization, Information Visualization and Visual Analytics.

Mike Kirby
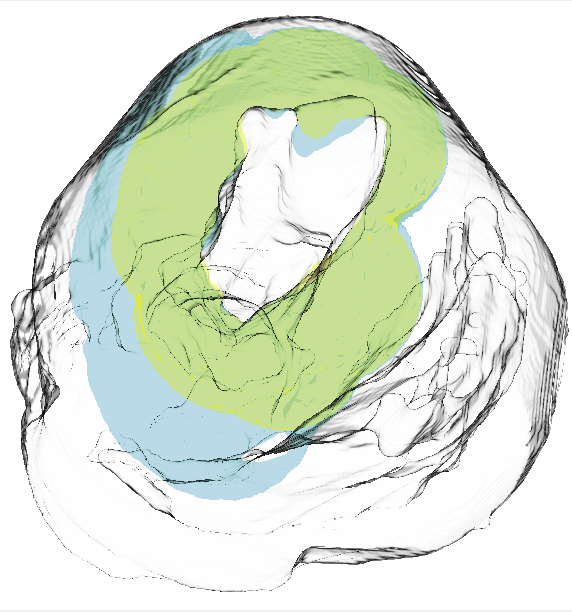
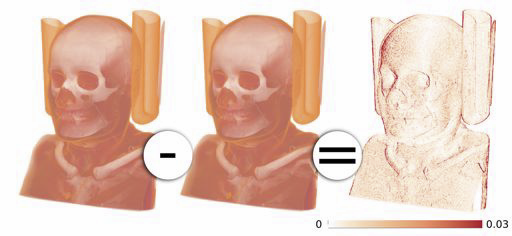
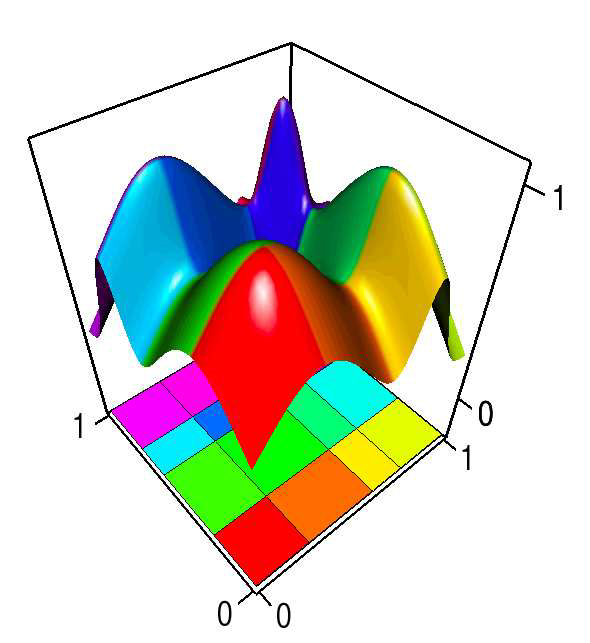
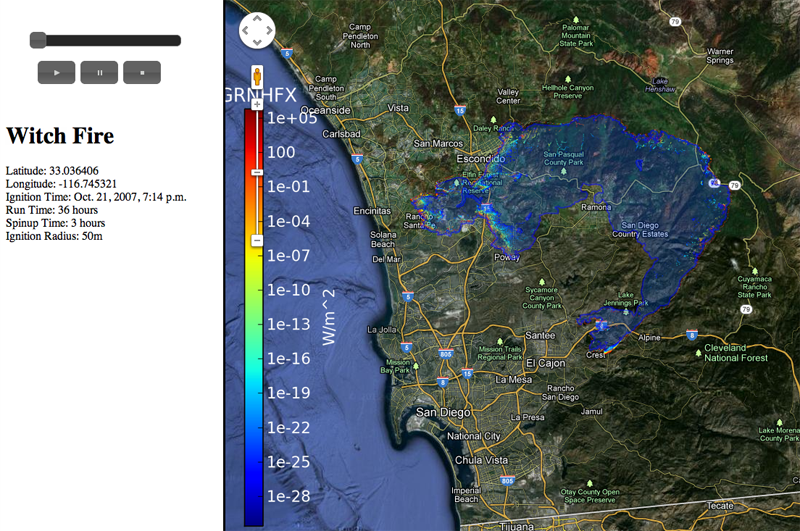
Uncertainty Visualization
Alex Lex
Information VisualizationCenters and Labs:
- Visualization Design Lab (VDL)
- CEDMAV
- POWDER Display Wall
- Modeling, Display, and Understanding Uncertainty in Simulations for Policy Decision Making
- Topological Data Analysis for Large Network Visualization
Funded Research Projects:
Publications in Visualization:
  Morse-Smale Regression S. Gerber, O. Reubel, P.-T. Bremer, V. Pascucci, R.T. Whitaker. In Journal of Computational and Graphical Statistics, Vol. 22, No. 1, pp. 193--214. 2013. DOI: 10.1080/10618600.2012.657132 This paper introduces a novel partition-based regression approach that incorporates topological information. Partition-based regression typically introduce a quality-of-fit-driven decomposition of the domain. The emphasis in this work is on a topologically meaningful segmentation. Thus, the proposed regression approach is based on a segmentation induced by a discrete approximation of the Morse-Smale complex. This yields a segmentation with partitions corresponding to regions of the function with a single minimum and maximum that are often well approximated by a linear model. This approach yields regression models that are amenable to interpretation and have good predictive capacity. Typically, regression estimates are quantified by their geometrical accuracy. For the proposed regression, an important aspect is the quality of the segmentation itself. Thus, this paper introduces a new criterion that measures the topological accuracy of the estimate. The topological accuracy provides a complementary measure to the classical geometrical error measures and is very sensitive to over-fitting. The Morse-Smale regression is compared to state-of-the-art approaches in terms of geometry and topology and yields comparable or improved fits in many cases. Finally, a detailed study on climate-simulation data demonstrates the application of the Morse-Smale regression. Supplementary materials are available online and contain an implementation of the proposed approach in the R package msr, an analysis and simulations on the stability of the Morse-Smale complex approximation and additional tables for the climate-simulation study. |
  Comments on the “Meshless Helmholtz-Hodge decomposition” H. Bhatia, G. Norgard, V. Pascucci, P.-T. Bremer. In IEEE Transactions on Visualization and Computer Graphics, Vol. 19, No. 3, pp. 527--528. 2013. DOI: 10.1109/TVCG.2012.62 The Helmholtz-Hodge decomposition (HHD) is one of the fundamental theorems of fluids describing the decomposition of a flow field into its divergence-free, curl-free and harmonic components. Solving for an HDD is intimately connected to the choice of boundary conditions which determine the uniqueness and orthogonality of the decomposition. This article points out that one of the boundary conditions used in a recent paper \"Meshless Helmholtz-Hodge decomposition\" [5] is, in general, invalid and provides an analytical example demonstrating the problem. We hope that this clarification on the theory will foster further research in this area and prevent undue problems in applying and extending the original approach. |
  Visualizing Invariant Manifolds in Area-Preserving Maps X. Tricoche, C. Garth, A. Sanderson, K. Joy. In Topological Methods in Data Analysis and Visualization II: Theory, Algorithms, and Applications, Edited by R. Peikert, H. Hauser, H. Carr, R. Fuchs, Springer Berlin Heidelberg, pp. 109--124. 2012. ISBN: 978-3-642-23175-9 DOI: 10.1007/978-3-642-23175-9_8 Area-preserving maps arise in the study of conservative dynamical systems describing a wide variety of physical phenomena, from the rotation of planets to the dynamics of a fluid. The visual inspection of these maps reveals a remarkable topological picture in which invariant manifolds form the fractal geometric scaffold of both quasi-periodic and chaotic regions. We discuss in this paper the visualization of such maps built upon these invariant manifolds. This approach is in stark contrast with the discrete Poincare plots that are typically used for the visual inspection of maps. We propose to that end several modified definitions of the finite-time Lyapunov exponents that we apply to reveal the underlying structure of the dynamics. We examine the impact of various parameters and the numerical aspects that pertain to the implementation of this method. We apply our technique to a standard analytical example and to a numerical simulation of magnetic confinement in a fusion reactor. In both cases our simple method is able to reveal salient structures across spatial scales and to yield expressive images across application domains. |
  enRoute: Dynamic Path Extraction from Biological Pathway Maps for In-Depth Experimental Data Analysis C Partl, A Lex, M Streit, D Kalkofen, K Kashofer, D Schmalstieg. In Proceedings of the IEEE Symposium on Biological Data Visualization (BioVis '12), IEEE, pp. 107--114. 2012. DOI: 10.1109/BioVis.2012.6378600 Pathway maps are an important source of information when analyzing functional implications of experimental data on biological processes. However, associating large quantities of data with nodes on a pathway map and allowing in depth-analysis at the same time is a challenging task. While a wide variety of approaches for doing so exist, they either do not scale beyond a few experiments or fail to represent the pathway appropriately. To remedy this, we introduce enRoute, a new approach for interactively exploring experimental data along paths that are dynamically extracted from pathways. By showing an extracted path side-by-side with experimental data, enRoute can present large amounts of data for every pathway node. It can visualize hundreds of samples, dozens of experimental conditions, and even multiple datasets capturing different aspects of a node at the same time. Another important property of this approach is its conceptual compatibility with arbitrary forms of pathways. Most notably, enRoute works well with pathways that are manually created, as they are available in large, public pathway databases. We demonstrate enRoute with pathways from the well-established KEGG database and expression as well as copy number datasets from humans and mice with more than 1,000 experiments. We validate enRoute using case studies with domain experts, who used enRoute to explore data for glioblastoma multiforme in humans and a model of steatohepatitis in mice. |
  StratomeX: Visual Analysis of Large-Scale Heterogeneous Genomics Data for Cancer Subtype Characterization A. Lex, M. Streit, H. Schulz, C. Partl, D. Schmalstieg, P.. Park, N. Gehlenborg. In Computer Graphics Forum (EuroVis '12), Vol. 31, No. 3, pp. 1175--1184. 2012. ISSN: 0167-7055 DOI: 10.1111/j.1467-8659.2012.03110.x dentification and characterization of cancer subtypes are important areas of research that are based on the integrated analysis of multiple heterogeneous genomics datasets. Since there are no tools supporting this process, much of this work is done using ad-hoc scripts and static plots, which is inefficient and limits visual exploration of the data. To address this, we have developed StratomeX, an integrative visualization tool that allows investigators to explore the relationships of candidate subtypes across multiple genomic data types such as gene expression, DNA methylation, or copy number data. StratomeX represents datasets as columns and subtypes as bricks in these columns. Ribbons between the columns connect bricks to show subtype relationships across datasets. Drill-down features enable detailed exploration. StratomeX provides insights into the functional and clinical implications of candidate subtypes by employing small multiples, which allow investigators to assess the effect of subtypes on molecular pathways or outcomes such as patient survival. As the configuration of viewing parameters in such a multi-dataset, multi-view scenario is complex, we propose a meta visualization and configuration interface for dataset dependencies and data-view relationships. StratomeX is developed in close collaboration with domain experts. We describe case studies that illustrate how investigators used the tool to explore subtypes in large datasets and demonstrate how they efficiently replicated findings from the literature and gained new insights into the data. |
  Visualizing Uncertainty in Biological Expression Data C. Holzhüter, A. Lex, D. Schmalstieg, H. Schulz, H. Schumann, M. Streit. In Proceedings of the SPIE Conference on Visualization and Data Analysis (VDA '12), Vol. 8294, pp. 82940O-82940O-11. 2012. DOI: 10.1117/12.908516 Expression analysis of ~omics data using microarrays has become a standard procedure in the life sciences. However, microarrays are subject to technical limitations and errors, which render the data gathered likely to be uncertain. While a number of approaches exist to target this uncertainty statistically, it is hardly ever even shown when the data is visualized using for example clustered heatmaps. Yet, this is highly useful when trying not to omit data that is "good enough" for an analysis, which otherwise would be discarded as too unreliable by established conservative thresholds. Our approach addresses this shortcoming by first identifying the margin above the error threshold of uncertain, yet possibly still useful data. It then displays this uncertain data in the context of the valid data by enhancing a clustered heatmap. We employ different visual representations for the different kinds of uncertainty involved. Finally, it lets the user interactively adjust the thresholds, giving visual feedback in the heatmap representation, so that an informed choice on which thresholds to use can be made instead of applying the usual rule-of-thumb cut-offs. We exemplify the usefulness of our concept by giving details for a concrete use case from our partners at the Medical University of Graz, thereby demonstrating our implementation of the general approach. |
  Model-Driven Design for the Visual Analysis of Heterogeneous Data M. Streit, H. Schulz, A. Lex, D. Schmalstieg, H. Schumann. In IEEE Transactions on Visualization and Computer Graphics, Vol. 18, No. 6, IEEE, pp. 998--1010. 2012. DOI: 10.1109/TVCG.2011.108 As heterogeneous data from different sources are being increasingly linked, it becomes difficult for users to understand how the data are connected, to identify what means are suitable to analyze a given data set, or to find out how to proceed for a given analysis task. We target this challenge with a new model-driven design process that effectively codesigns aspects of data, view, analytics, and tasks. We achieve this by using the workflow of the analysis task as a trajectory through data, interactive views, and analytical processes. The benefits for the analysis session go well beyond the pure selection of appropriate data sets and range from providing orientation or even guidance along a preferred analysis path to a potential overall speedup, allowing data to be fetched ahead of time. We illustrate the design process for a biomedical use case that aims at determining a treatment plan for cancer patients from the visual analysis of a large, heterogeneous clinical data pool. As an example for how to apply the comprehensive design approach, we present Stack'n'flip, a sample implementation which tightly integrates visualizations of the actual data with a map of available data sets, views, and tasks, thus capturing and communicating the analytical workflow through the required data sets. |
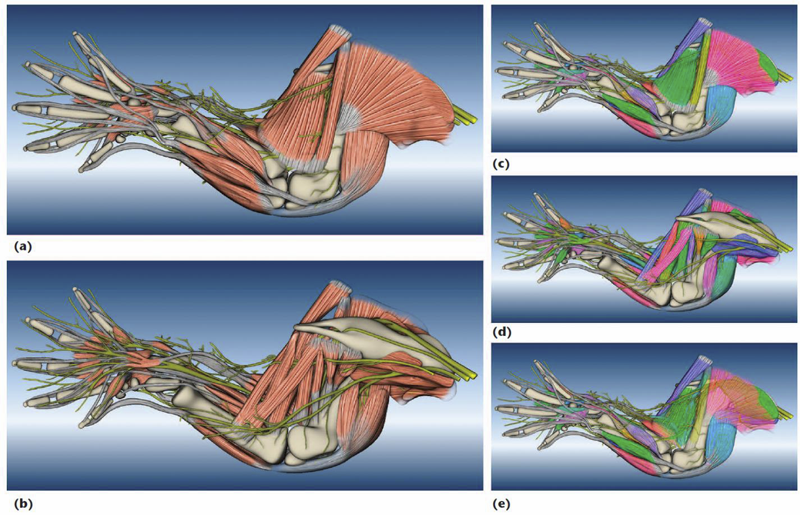
  A Practical Workflow for Making Anatomical Atlases in Biological Research Y. Wan, A.K. Lewis, M. Colasanto, M. van Langeveld, G. Kardon, C.D. Hansen. In IEEE Computer Graphics and Applications, Vol. 32, No. 5, pp. 70--80. 2012. DOI: 10.1109/MCG.2012.64 An anatomical atlas provides a detailed map for medical and biological studies of anatomy. These atlases are important for understanding normal anatomy and the development and function of structures, and for determining the etiology of congenital abnormalities. Unfortunately, for biologists, generating such atlases is difficult, especially ones with the informative content and aesthetic quality that characterize human anatomy atlases. Building such atlases requires knowledge of the species being studied and experience with an art form that can faithfully record and present this knowledge, both of which require extensive training in considerably different fields. (For some background on anatomical atlases, see the related sidebar.) With the latest innovations in data acquisition and computing techniques, atlas building has changed dramatically. We can now create atlases from 3D images of biological specimens, allowing for high-quality, faithful representations. Labeling of structures using fluorescently tagged antibodies, confocal 3D scanning of these labeled structures, volume rendering, segmentation, and surface reconstruction techniques all promise solutions to the problem of building atlases. However, biology researchers still ask, \"Is there a set of tools we can use or a practical workflow we can follow so that we can easily build models from our biological data?\" To help answer this question, computer scientists have developed many algorithms, tools, and program codes. Unfortunately, most of these researchers have tackled only one aspect of the problem or provided solutions to special cases. So, the general question of how to build anatomical atlases remains unanswered. For a satisfactory answer, biologists need a practical workflow they can easily adapt for different applications. In addition, reliable tools that can fit into the workflow must be readily available. Finally, examples using the workflow and tools to build anatomical atlases would demonstrate these resources' utility for biological research. To build a mouse limb atlas for studying the development of the limb musculoskeletal system, University of Utah biologists, artists, and computer scientists have designed a generalized workflow for generating anatomical atlases. We adapted it from a CG artist's workflow of building 3D models for animated films and video games. The tools we used to build the atlas were mostly commercial, industry-standard software packages. Having been developed, tested, and employed for industrial use for decades, CG artists' workflow and tools, with certain adaptations, are the most suitable for making high-quality anatomical atlases, especially under strict budgetary and time limits. Biological researchers have been largely unaware of these resources. By describing our experiences in this project, we hope to show biologists how to use these resources to make anatomically accurate, high-quality, and useful anatomical atlases. |
  Aggregate Gaze Visualization with Real-Time Heatmaps A. Duchowski, M. Price, M.D. Meyer, P. Orero. In Proceedings of the ACM Symposium on Eye Tracking Research and Applications (ETRA), pp. 13--20. 2012. DOI: 10.1145/2168556.2168558 A GPU implementation is given for real-time visualization of aggregate eye movements (gaze) via heatmaps. Parallelization of the algorithm leads to substantial speedup over its CPU-based implementation and, for the first time, allows real-time rendering of heatmaps atop video. GLSL shader colorization allows the choice of color ramps. Several luminance-based color maps are advocated as alternatives to the popular rainbow color map, considered inappropriate (harmful) for depiction of (relative) gaze distributions. |
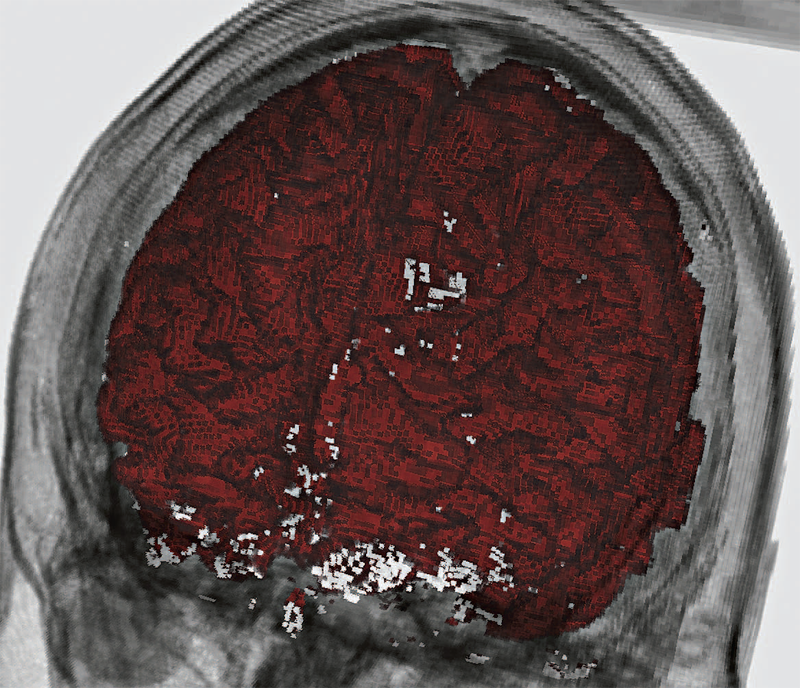
  Gaussian Mixture Model Based Volume Visualization S. Liu, J.A. Levine, P.-T. Bremer, V. Pascucci. In Proceedings of the IEEE Large-Scale Data Analysis and Visualization Symposium 2012, Note: Received Best Paper Award, pp. 73--77. 2012. DOI: 10.1109/LDAV.2012.6378978 Representing uncertainty when creating visualizations is becoming more indispensable to understand and analyze scientific data. Uncertainty may come from different sources, such as, ensembles of experiments or unavoidable information loss when performing data reduction. One natural model to represent uncertainty is to assume that each position in space instead of a single value may take on a distribution of values. In this paper we present a new volume rendering method using per voxel Gaussian mixture models (GMMs) as the input data representation. GMMs are an elegant and compact way to drastically reduce the amount of data stored while still enabling realtime data access and rendering on the GPU. Our renderer offers efficient sampling of the data distribution, generating renderings of the data that flicker at each frame to indicate high variance. We can accumulate samples as well to generate still frames of the data, which preserve additional details in the data as compared to either traditional scalar indicators (such as a mean or a single nearest neighbor down sample) or to fitting the data with only a single Gaussian per voxel. We demonstrate the effectiveness of our method using ensembles of climate simulations and MRI scans as well as the down sampling of large scalar fields as examples. Keywords: Uncertainty Visualization, Volume Rendering, Gaussian Mixture Model, Ensemble Visualization |