Image Analysis
SCI's imaging work addresses fundamental questions in 2D and 3D image processing, including filtering, segmentation, surface reconstruction, and shape analysis. In low-level image processing, this effort has produce new nonparametric methods for modeling image statistics, which have resulted in better algorithms for denoising and reconstruction. Work with particle systems has led to new methods for visualizing and analyzing 3D surfaces. Our work in image processing also includes applications of advanced computing to 3D images, which has resulted in new parallel algorithms and real-time implementations on graphics processing units (GPUs). Application areas include medical image analysis, biological image processing, defense, environmental monitoring, and oil and gas.
Ross Whitaker
Segmentation
Chris Johnson
Diffusion Tensor AnalysisFunded Research Projects:
Publications in Image Analysis:
  Manifold learning for analysis of low-order nonlinear dynamics in high-dimensional electrocardiographic signals B. Erem, P. Stovicek, D.H. Brooks. In Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 844--847. 2012. DOI: 10.1109/ISBI.2012.6235680 The dynamical structure of electrical recordings from the heart or torso surface is a valuable source of information about cardiac physiological behavior. In this paper, we use an existing data-driven technique for manifold identification to reveal electrophysiologically significant changes in the underlying dynamical structure of these signals. Our results suggest that this analysis tool characterizes and differentiates important parameters of cardiac bioelectric activity through their dynamic behavior, suggesting the potential to serve as an effective dynamic constraint in the context of inverse solutions. |
  Validation study of automated dermal/epidermal junction localization algorithm in reflectance confocal microscopy images of skin S. Kurugol, M. Rajadhyaksha, J.G. Dy, D.H. Brooks. In Proceedings of SPIE Photonic Therapeutics and Diagnostics VIII, Vol. 8207, No. 1, pp. 820702-820711. 2012. DOI: 10.1117/12.909227 PubMed ID: 24376908 PubMed Central ID: PMC3872972 Reflectance confocal microscopy (RCM) has seen increasing clinical application for noninvasive diagnosis of skin cancer. Identifying the location of the dermal-epidermal junction (DEJ) in the image stacks is key for effective clinical imaging. For example, one clinical imaging procedure acquires a dense stack of 0.5x0.5mm FOV images and then, after manual determination of DEJ depth, collects a 5x5mm mosaic at that depth for diagnosis. However, especially in lightly pigmented skin, RCM images have low contrast at the DEJ which makes repeatable, objective visual identification challenging. We have previously published proof of concept for an automated algorithm for DEJ detection in both highly- and lightly-pigmented skin types based on sequential feature segmentation and classification. In lightly-pigmented skin the change of skin texture with depth was detected by the algorithm and used to locate the DEJ. Here we report on further validation of our algorithm on a more extensive collection of 24 image stacks (15 fair skin, 9 dark skin). We compare algorithm performance against classification by three clinical experts. We also evaluate inter-expert consistency among the experts. The average correlation across experts was 0.81 for lightly pigmented skin, indicating the difficulty of the problem. The algorithm achieved epidermis/dermis misclassification rates smaller than 10% (based on 25x25 mm tiles) and average distance from the expert labeled boundaries of ~6.4 ?m for fair skin and ~5.3 ?m for dark skin, well within average cell size and less than 2x the instrument resolution in the optical axis. |
  Interactive Extraction of Neural Structures with User-Guided Morphological Diffusion Y. Wan, H. Otsuna, C.-B. Chien, C.D. Hansen. In Proceedings of the IEEE Symposium on Biological Data Visualization, pp. 1--8. 2012. DOI: 10.1109/BioVis.2012.6378577 Extracting neural structures with their fine details from confocal volumes is essential to quantitative analysis in neurobiology research. Despite the abundance of various segmentation methods and tools, for complex neural structures, both manual and semi-automatic methods are ineffective either in full 3D or when user interactions are restricted to 2D slices. Novel interaction techniques and fast algorithms are demanded by neurobiologists to interactively and intuitively extract neural structures from confocal data. In this paper, we present such an algorithm-technique combination, which lets users interactively select desired structures from visualization results instead of 2D slices. By integrating the segmentation functions with a confocal visualization tool neurobiologists can easily extract complex neural structures within their typical visualization workflow. |
  A Practical Workflow for Making Anatomical Atlases in Biological Research Y. Wan, A.K. Lewis, M. Colasanto, M. van Langeveld, G. Kardon, C.D. Hansen. In IEEE Computer Graphics and Applications, Vol. 32, No. 5, pp. 70--80. 2012. DOI: 10.1109/MCG.2012.64 An anatomical atlas provides a detailed map for medical and biological studies of anatomy. These atlases are important for understanding normal anatomy and the development and function of structures, and for determining the etiology of congenital abnormalities. Unfortunately, for biologists, generating such atlases is difficult, especially ones with the informative content and aesthetic quality that characterize human anatomy atlases. Building such atlases requires knowledge of the species being studied and experience with an art form that can faithfully record and present this knowledge, both of which require extensive training in considerably different fields. (For some background on anatomical atlases, see the related sidebar.) With the latest innovations in data acquisition and computing techniques, atlas building has changed dramatically. We can now create atlases from 3D images of biological specimens, allowing for high-quality, faithful representations. Labeling of structures using fluorescently tagged antibodies, confocal 3D scanning of these labeled structures, volume rendering, segmentation, and surface reconstruction techniques all promise solutions to the problem of building atlases. However, biology researchers still ask, \"Is there a set of tools we can use or a practical workflow we can follow so that we can easily build models from our biological data?\" To help answer this question, computer scientists have developed many algorithms, tools, and program codes. Unfortunately, most of these researchers have tackled only one aspect of the problem or provided solutions to special cases. So, the general question of how to build anatomical atlases remains unanswered. For a satisfactory answer, biologists need a practical workflow they can easily adapt for different applications. In addition, reliable tools that can fit into the workflow must be readily available. Finally, examples using the workflow and tools to build anatomical atlases would demonstrate these resources' utility for biological research. To build a mouse limb atlas for studying the development of the limb musculoskeletal system, University of Utah biologists, artists, and computer scientists have designed a generalized workflow for generating anatomical atlases. We adapted it from a CG artist's workflow of building 3D models for animated films and video games. The tools we used to build the atlas were mostly commercial, industry-standard software packages. Having been developed, tested, and employed for industrial use for decades, CG artists' workflow and tools, with certain adaptations, are the most suitable for making high-quality anatomical atlases, especially under strict budgetary and time limits. Biological researchers have been largely unaware of these resources. By describing our experiences in this project, we hope to show biologists how to use these resources to make anatomically accurate, high-quality, and useful anatomical atlases. |
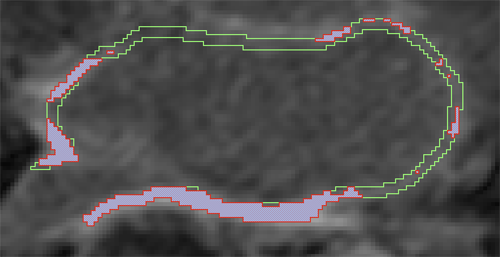
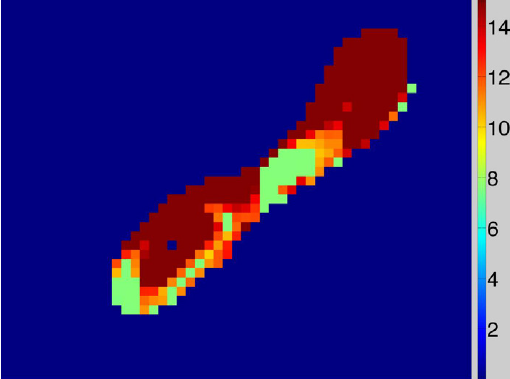
  Automatic classification of scar tissue in late gadolinium enhancement cardiac MRI for the assessment of left-atrial wall injury after radiofrequency ablation D. Perry, A. Morris, N. Burgon, C. McGann, R.S. MacLeod, J. Cates. In SPIE Proceedings, Vol. 8315, pp. (published online). 2012. DOI: 10.1117/12.910833 PubMed ID: 24236224 PubMed Central ID: PMC3824273 Radiofrequency ablation is a promising procedure for treating atrial fibrillation (AF) that relies on accurate lesion delivery in the left atrial (LA) wall for success. Late Gadolinium Enhancement MRI (LGE MRI) at three months post-ablation has proven effective for noninvasive assessment of the location and extent of scar formation, which are important factors for predicting patient outcome and planning of redo ablation procedures. We have developed an algorithm for automatic classification in LGE MRI of scar tissue in the LA wall and have evaluated accuracy and consistency compared to manual scar classifications by expert observers. Our approach clusters voxels based on normalized intensity and was chosen through a systematic comparison of the performance of multivariate clustering on many combinations of image texture. Algorithm performance was determined by overlap with ground truth, using multiple overlap measures, and the accuracy of the estimation of the total amount of scar in the LA. Ground truth was determined using the STAPLE algorithm, which produces a probabilistic estimate of the true scar classification from multiple expert manual segmentations. Evaluation of the ground truth data set was based on both inter- and intra-observer agreement, with variation among expert classifiers indicating the difficulty of scar classification for a given a dataset. Our proposed automatic scar classification algorithm performs well for both scar localization and estimation of scar volume: for ground truth datasets considered easy, variability from the ground truth was low; for those considered difficult, variability from ground truth was on par with the variability across experts. |
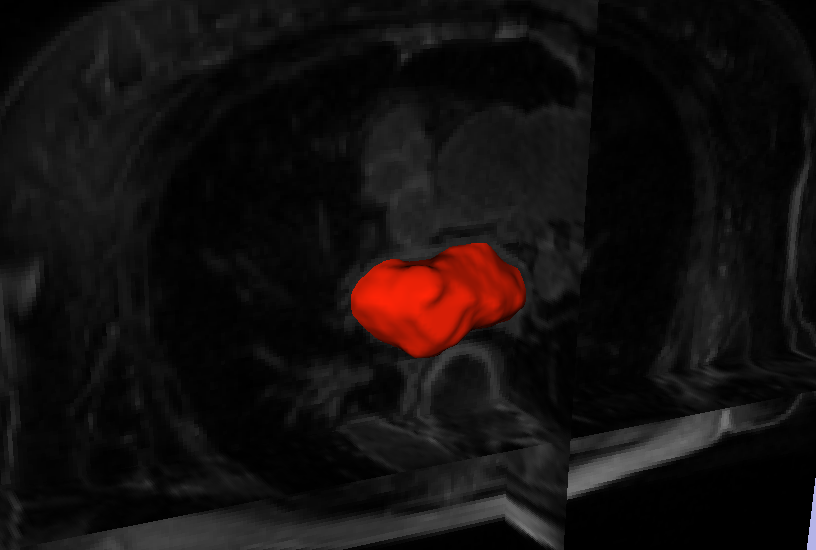
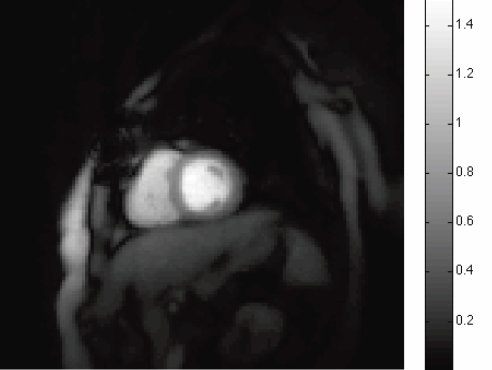
  Automatic Segmentation of the Left Atrium from MRI Images using Salient Feature and Contour Evolution L. Zhu, Y. Gao, A. Yezzi, R.S. MacLeod, J. Cates, A. Tannenbaum. In Proceedings of the 34th Annual International Conference of the IEEE EMBS, pp. 3211--214. 2012. DOI: 10.1109/EMBC.2012.6346648 PubMed ID: 23366609 PubMed Central ID: PMC3652873 We propose an automatic approach for segmenting the left atrium from MRI images. In particular, the thoracic aorta is detected and used as a salient feature to find a seed region that lies inside the left atrium. A hybrid energy that combines robust statistics and localized region intensity information is employed to evolve active contours from the seed region to capture the whole left atrium. The experimental results demonstrate the accuracy and robustness of our approach. |
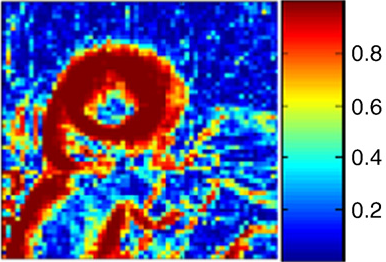
  Fingerprint Image Segmentation using Data Manifold Characteristic Features A.R.C. Paiva, T. Tasdizen. In International Journal of Pattern Recognition and Artificial Intelligence, Vol. 26, No. 4, pp. (23 pages). 2012. DOI: 10.1142/S0218001412560101 Automatic fingerprint identification systems (AFIS) have been studied extensively and are widely used for biometric identification. Given its importance, many well-engineered methods have been developed for the different stages that encompass those systems. The first stage of any such system is the segmentation of the actual fingerprint region from the background. This is typically achieved by classifying pixels, or blocks of pixels, based on a set of features. In this paper, we describe novel features for fingerprint segmentation that express the underlying manifold topology associated with image patches in a local neighborhood. It is shown that fingerprint patches seen in a high-dimensional space form a simple and highly regular circular manifold. The characterization of the manifold topology suggests a set of optimal features that characterize the local properties of the fingerprint. Thus, fingerprint segmentation can be formulated as a classification problem based on the deviation from the expected topology. This leads to features that are more robust to changes in contrast than mean, variance and coherence. The superior performance of the proposed features for fingerprint segmentation is shown in the eight datasets from the 2002 and 2004 Fingerprint Verification Competitions. Keywords: Fingerprint segmentation, manifold characterization, feature extraction, dimensionality reduction |
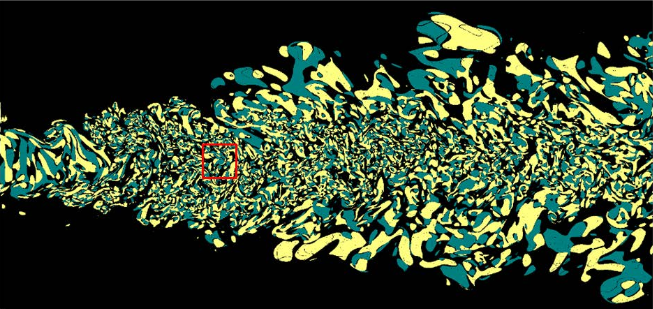
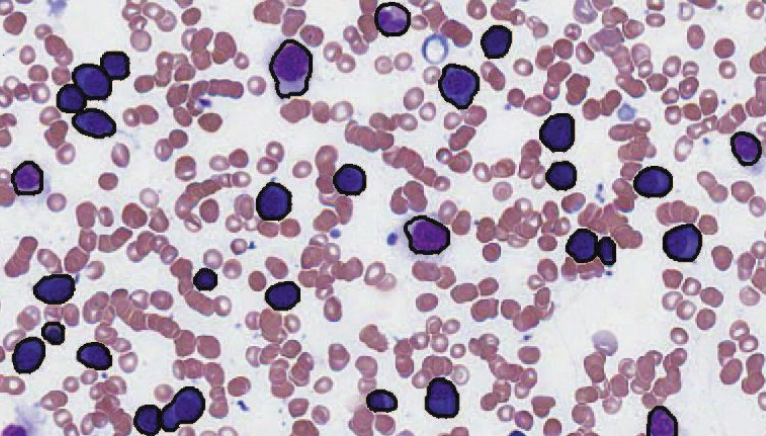
  Segmentation of Haematopoeitic Cells in Bone Marrow Using Circle Detection and Splitting Techniques N. Ramesh, M.E. Salama, T. Tasdizen. In 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 206--209. 2012. DOI: 10.1109/ISBI.2012.6235520 Bone marrow evaluation is indicated when peripheral blood abnormalities are not explained by clinical, physical, or laboratory findings. In this paper, we propose a novel method for segmentation of haematopoietic cells in the bone marrow from scanned slide images. Segmentation of clumped cells is a challenging problem for this application. We first use color information and morphology to eliminate red blood cells and the background. Clumped haematopoietic cells are then segmented using circle detection and a splitting algorithm based on the detected circle centers. The Hough Transform is used for circle detection and to find the number and positions of circle centers in each region. The splitting algorithm is based on detecting the maximum curvature points, and partitioning them based on information obtained from the centers of the circles in each region. The performance of the segmentation algorithm for haematopoietic cells is evaluated by comparing our proposed method with a hematologist's visual segmentation in a set of 3748 cells. |
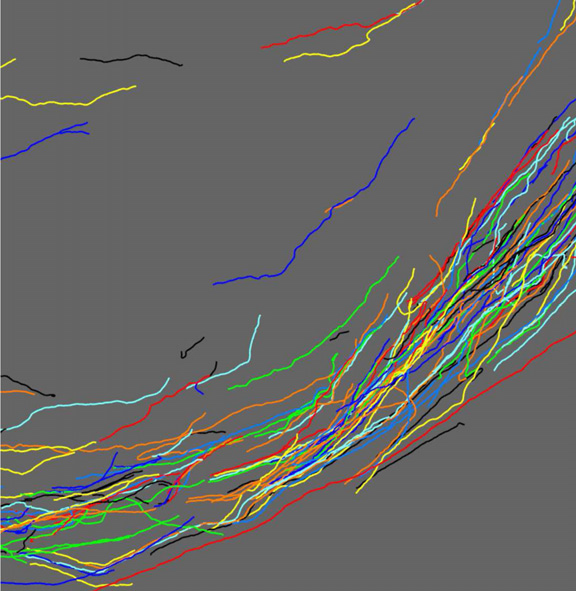
  Serial section registration of axonal confocal microscopy datasets for long-range neural circuit reconstruction L. Hogrebe, A.R.C. Paiva, E. Jurrus, C. Christensen, M. Bridge, L. Dai, R.L. Pfeiffer, P.R. Hof, B. Roysam, J.R. Korenberg, T. Tasdizen. In Journal of Neuroscience Methods, Vol. 207, No. 2, pp. 200--210. 2012. DOI: 10.1016/j.jneumeth.2012.03.002 In the context of long-range digital neural circuit reconstruction, this paper investigates an approach for registering axons across histological serial sections. Tracing distinctly labeled axons over large distances allows neuroscientists to study very explicit relationships between the brain's complex interconnects and, for example, diseases or aberrant development. Large scale histological analysis requires, however, that the tissue be cut into sections. In immunohistochemical studies thin sections are easily distorted due to the cutting, preparation, and slide mounting processes. In this work we target the registration of thin serial sections containing axons. Sections are first traced to extract axon centerlines, and these traces are used to define registration landmarks where they intersect section boundaries. The trace data also provides distinguishing information regarding an axon's size and orientation within a section. We propose the use of these features when pairing axons across sections in addition to utilizing the spatial relationships among the landmarks. The global rotation and translation of an unregistered section are accounted for using a random sample consensus (RANSAC) based technique. An iterative nonrigid refinement process using B-spline warping is then used to reconnect axons and produce the sought after connectivity information. |
  Edge enhanced spatio-temporal constrained reconstruction of undersampled dynamic contrast enhanced radial MRI E.V.R. DiBella, T. Tasdizen. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 704--707. 2012. DOI: 10.1109/ISBI.2010.5490077 There are many applications in MRI where it is desirable to have high spatial and high temporal resolution. This can be achieved by undersampling of k-space and requires special techniques for reconstruction. Even if undersampling artifacts are removed, sharpness of the edges can be a problem. We propose a new technique that uses the gradient from a reference image to improve the quality of the edges in the reconstructed image along with a spatio-temporal constraint to reduce aliasing artifacts and noise. The reference is created from undersampled dynamic data by combining several adjacent frames. The method was tested on undersampled radial DCE MRI data with little respiratory motion. The proposed method was compared to reconstruction using the spatio-temporal constrained reconstruction. Sharper edges and an increase in the contrast was observed by using the proposed method. |
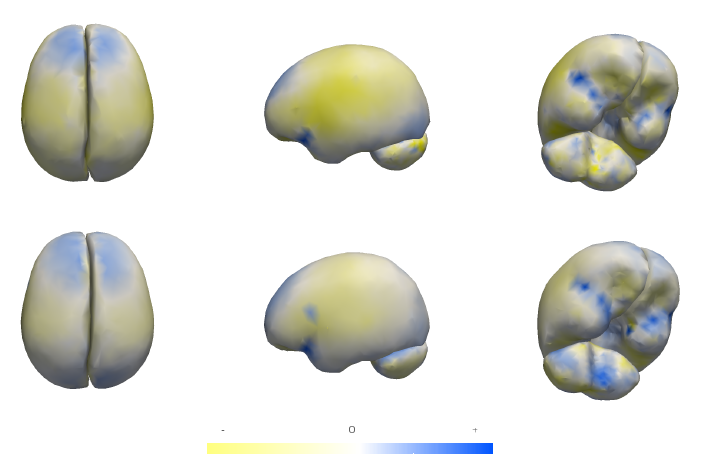
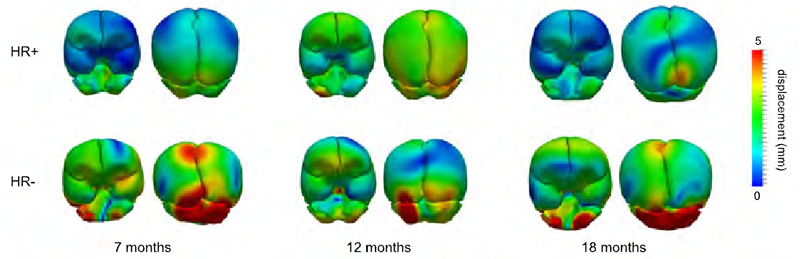
  Differences in subcortical structures in young adolescents at familial risk for schizophrenia: A preliminary study M.K. Dougherty, H. Gu, J. Bizzell, S. Ramsey, G. Gerig, D.O. Perkins, A. Belger. In Psychiatry Res., pp. (Epub ahead of print. Nov. 9, 2012. DOI: 10.1016/j.pscychresns.2012.04.016 PubMed ID: 23146250 |
  How Many Templates Does It Take for a Good Segmentation?: Error Analysis in Multiatlas Segmentation as a Function of Database Size S.P. Awate, P. Zhu, R.T. Whitaker. In Int. Workshop Multimodal Brain Image Analysis (MBIA) at Int. Conf. MICCAI, Lecture Notes in Computer Science (LNCS), Vol. 2, Note: Recieved Best Paper Award, pp. 103--114. 2012. PubMed ID: 24501720 PubMed Central ID: PMC3910563 This paper proposes a novel formulation to model and analyze the statistical characteristics of some types of segmentation problems that are based on combining label maps / templates / atlases. Such segmentation-by-example approaches are quite powerful on their own for several clinical applications and they provide prior information, through spatial context, when combined with intensity-based segmentation methods. The proposed formulation models a class of multiatlas segmentation problems as nonparametric regression problems in the high-dimensional space of images. The paper presents a systematic analysis of the nonparametric estimation's convergence behavior (i.e. characterizing segmentation error as a function of the size of the multiatlas database) and shows that it has a specific analytic form involving several parameters that are fundamental to the specific segmentation problem (i.e. chosen anatomical structure, imaging modality, registration method, label-fusion algorithm, etc.). We describe how to estimate these parameters and show that several brain anatomical structures exhibit the trends determined analytically. The proposed framework also provides per-voxel confidence measures for the segmentation. We show that the segmentation error for large database sizes can be predicted using small-sized databases. Thus, small databases can be exploited to predict the database sizes required (\"how many templates\") to achieve \"good\" segmentations having errors lower than a specified tolerance. Such cost-benefit analysis is crucial for designing and deploying multiatlas segmentation systems. |
  Mixed-Effects Shape Models for Estimating Longitudinal Changes in Anatomy M. Datar, P. Muralidharan, A. Kumar, S. Gouttard, J. Piven, G. Gerig, R.T. Whitaker, P.T. Fletcher. In Spatio-temporal Image Analysis for Longitudinal and Time-Series Image Data, Lecture Notes in Computer Science, Vol. 7570, Springer Berlin / Heidelberg, pp. 76--87. 2012. ISBN: 978-3-642-33554-9 DOI: 10.1007/978-3-642-33555-6_7 In this paper, we propose a new method for longitudinal shape analysis that ts a linear mixed-e ects model, while simultaneously optimizing correspondences on a set of anatomical shapes. Shape changes are modeled in a hierarchical fashion, with the global population trend as a xed e ect and individual trends as random e ects. The statistical signi cance of the estimated trends are evaluated using speci cally designed permutation tests. We also develop a permutation test based on the Hotelling T2 statistic to compare the average shapes trends between two populations. We demonstrate the bene ts of our method on a synthetic example of longitudinal tori and data from a developmental neuroimaging study. Keywords: Computer Science |
  Analysis of Longitudinal Shape Variability via Subject Specific Growth Modeling J. Fishbaugh, M.W. Prastawa, S. Durrleman, G. Gerig. In Medical Image Computing and Computer-Assisted Intervention – Proceedings of MICCAI 2012, Lecture Notes in Computer Science (LNCS), Vol. 7510, pp. 731--738. October, 2012. DOI: 10.1007/978-3-642-33415-3_90 Statistical analysis of longitudinal imaging data is crucial for understanding normal anatomical development as well as disease progression. This fundamental task is challenging due to the difficulty in modeling longitudinal changes, such as growth, and comparing changes across different populations. We propose a new approach for analyzing shape variability over time, and for quantifying spatiotemporal population differences. Our approach estimates 4D anatomical growth models for a reference population (an average model) and for individuals in different groups. We define a reference 4D space for our analysis as the average population model and measure shape variability through diffeomorphisms that map the reference to the individuals. Conducting our analysis on this 4D space enables straightforward statistical analysis of deformations as they are parameterized by momenta vectors that are located at homologous locations in space and time. We evaluate our method on a synthetic shape database and clinical data from a study that seeks to quantify growth differences in subjects at risk for autism. |
  Neuroimaging of Structural Pathology and Connectomics in Traumatic Brain Injury: Toward Personalized Outcome Prediction A. Irimia, Bo Wang, S.R. Aylward, M.W. Prastawa, D.F. Pace, G. Gerig, D.A. Hovda, R.Kikinis, P.M. Vespa, J.D. Van Horn. In NeuroImage: Clinical, Vol. 1, No. 1, Elsvier, pp. 1--17. 2012. DOI: 10.1016/j.nicl.2012.08.002 Recent contributions to the body of knowledge on traumatic brain injury (TBI) favor the view that multimodal neuroimaging using structural and functional magnetic resonance imaging (MRI and fMRI, respectively) as well as diffusion tensor imaging (DTI) has excellent potential to identify novel biomarkers and predictors of TBI outcome. This is particularly the case when such methods are appropriately combined with volumetric/morphometric analysis of brain structures and with the exploration of TBI]related changes in brain network properties at the level of the connectome. In this context, our present review summarizes recent developments on the roles of these two techniques in the search for novel structural neuroimaging biomarkers that have TBI outcome prognostication value. The themes being explored cover notable trends in this area of research, including (1) the role of advanced MRI processing methods in the analysis of structural pathology, (2) the use of brain connectomics and network analysis to identify outcome biomarkers, and (3) the application of multivariate statistics to predict outcome using neuroimaging metrics. The goal of the review is to draw the communityfs attention to these recent advances on TBI outcome prediction methods and to encourage the development of new methodologies whereby structural neuroimaging can be used to identify biomarkers of TBI outcome. |