Image Analysis
SCI's imaging work addresses fundamental questions in 2D and 3D image processing, including filtering, segmentation, surface reconstruction, and shape analysis. In low-level image processing, this effort has produce new nonparametric methods for modeling image statistics, which have resulted in better algorithms for denoising and reconstruction. Work with particle systems has led to new methods for visualizing and analyzing 3D surfaces. Our work in image processing also includes applications of advanced computing to 3D images, which has resulted in new parallel algorithms and real-time implementations on graphics processing units (GPUs). Application areas include medical image analysis, biological image processing, defense, environmental monitoring, and oil and gas.
Ross Whitaker
Segmentation
Chris Johnson
Diffusion Tensor AnalysisFunded Research Projects:
Publications in Image Analysis:
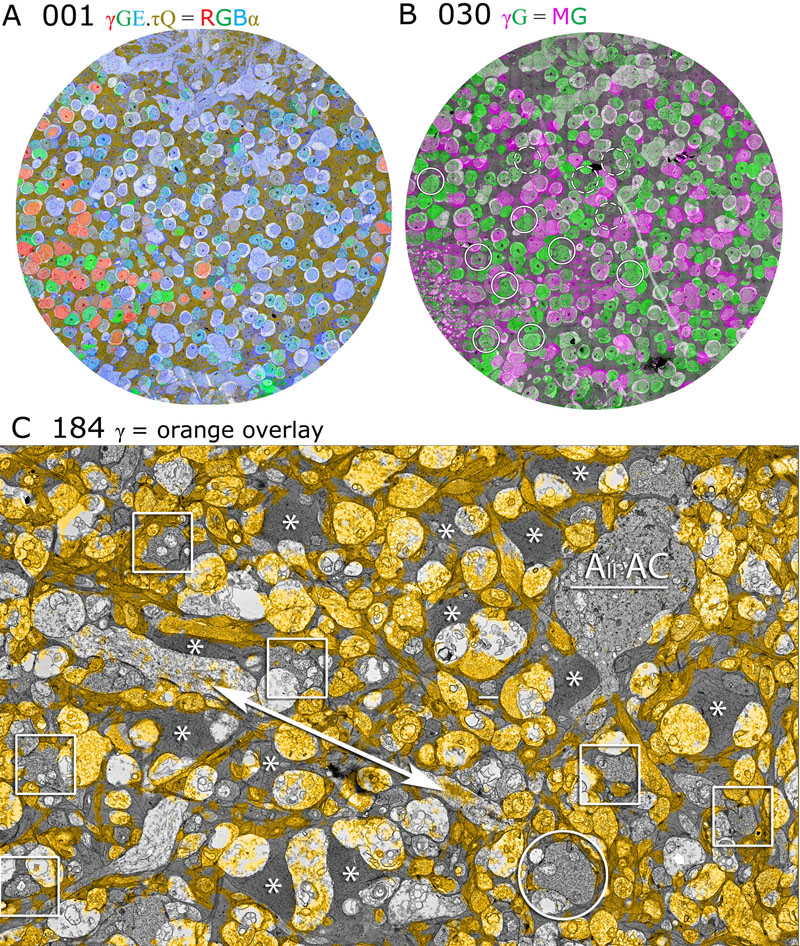
  Exploring the Retinal Connectome J.R. Anderson, B.W. Jones, C.B. Watt, M.V. Shaw, J.-H. Yang, D. DeMill, J.S. Lauritzen, Y. Lin, K.D. Rapp, D. Mastronarde, P. Koshevoy, B. Grimm, T. Tasdizen, R.T. Whitaker, R.E. Marc. In Molecular Vision, Vol. 17, pp. 355--379. 2011. PubMed ID: 21311605 Purpose: A connectome is a comprehensive description of synaptic connectivity for a neural domain. Our goal was to produce a connectome data set for the inner plexiform layer of the mammalian retina. This paper describes our first retinal connectome, validates the method, and provides key initial findings. Methods: We acquired and assembled a 16.5 terabyte connectome data set RC1 for the rabbit retina at .2 nm resolution using automated transmission electron microscope imaging, automated mosaicking, and automated volume registration. RC1 represents a column of tissue 0.25 mm in diameter, spanning the inner nuclear, inner plexiform, and ganglion cell layers. To enhance ultrastructural tracing, we included molecular markers for 4-aminobutyrate (GABA), glutamate, glycine, taurine, glutamine, and the in vivo activity marker, 1-amino-4-guanidobutane. This enabled us to distinguish GABAergic and glycinergic amacrine cells; to identify ON bipolar cells coupled to glycinergic cells; and to discriminate different kinds of bipolar, amacrine, and ganglion cells based on their molecular signatures and activity. The data set was explored and annotated with Viking, our multiuser navigation tool. Annotations were exported to additional applications to render cells, visualize network graphs, and query the database. Results: Exploration of RC1 showed that the 2 nm resolution readily recapitulated well known connections and revealed several new features of retinal organization: (1) The well known AII amacrine cell pathway displayed more complexity than previously reported, with no less than 17 distinct signaling modes, including ribbon synapse inputs from OFF bipolar cells, wide-field ON cone bipolar cells and rod bipolar cells, and extensive input from cone-pathway amacrine cells. (2) The axons of most cone bipolar cells formed a distinct signal integration compartment, with ON cone bipolar cell axonal synapses targeting diverse cell types. Both ON and OFF bipolar cells receive axonal veto synapses. (3) Chains of conventional synapses were very common, with intercalated glycinergic-GABAergic chains and very long chains associated with starburst amacrine cells. Glycinergic amacrine cells clearly play a major role in ON-OFF crossover inhibition. (4) Molecular and excitation mapping clearly segregates ultrastructurally defined bipolar cell groups into different response clusters. (5) Finally, low-resolution electron or optical imaging cannot reliably map synaptic connections by process geometry, as adjacency without synaptic contact is abundant in the retina. Only direct visualization of synapses and gap junctions suffices. Conclusions: Connectome assembly and analysis using conventional transmission electron microscopy is now practical for network discovery. Our surveys of volume RC1 demonstrate that previously studied systems such as the AII amacrine cell network involve more network motifs than previously known. The AII network, primarily considered a scotopic pathway, clearly derives ribbon synapse input from photopic ON and OFF cone bipolar cell networks and extensive photopic GABAergic amacrine cell inputs. Further, bipolar cells show extensive inputs and outputs along their axons, similar to multistratified nonmammalian bipolar cells. Physiologic evidence of significant ON-OFF channel crossover is strongly supported by our anatomic data, showing alternating glycine-to-GABA paths. Long chains of amacrine cell networks likely arise from homocellular GABAergic synapses between starburst amacrine cells. Deeper analysis of RC1 offers the opportunity for more complete descriptions of specific networks. Keywords: neuroscience, retina, vision, blindness, visus, crcns |
  Trace Driven Registration of Neuron Confocal Microscopy Stacks L. Hogrebe, A. Paiva, E. Jurrus, C. Christensen, M. Bridge, J.R. Korenberg, T. Tasdizen. In IEEE International Symposium on Biomedical Imaging (ISBI), pp. 1345--1348. 2011. DOI: 10.1109/ISBI.2011.5872649 |
  The Viking Viewer: Scalable Multiuser Annotation and Summarization of Large Volume Datasets J.R. Anderson, B.C. Grimm, S. Mohammed, B.W. Jones, T. Tasdizen, J. Spaltenstein, P. Koshevoy, R.T. Whitaker, R.E. Marc. In Journal of Microscopy, Vol. 241, No. 1, pp. 13--28. 2010. DOI: 10.1111/j.1365-2818.2010.03402.x |
  Edge enhanced spatio-temporal constrained reconstruction of undersampled dynamic contrast enhanced radial MRI S.K. Iyer, E. DiBella, T. Tasdizen. In IEEE International Symposium on Biomedical Imaging (ISBI): From Nano to Macro, pp. 704--707. 2010. DOI: 10.1109/ISBI.2010.5490077 |
  Reconstruction of 3D Dynamic Contrast-Enhanced Magnetic Resonance Imaging Using Nonlocal Means G. Adluru, T. Tasdizen, M.C. Schabel, E.V.R. DiBella. In Journal of Magnetic Resonance Imaging, Vol. 32, pp. 1217--1227. 2010. DOI: 10.1002/jmri.22358 |
  Improving Undersampled MRI Reconstruction Using Non-Local Means G. Adluru, T. Tasdizen, R. Whitaker, E. DiBella. In Proceedings of the 2010 International Conference on Pattern Recognition, pp. 4000--4003. 2010. DOI: 10.1109/ICPR.2010.973 |
  Brain volumes in psychotic youth with schizophrenia and mood disorders M. El-Sayed, R.G. Steen, M.D. Poe, T.C. Bethea, G. Gerig, J. Lieberman, L. Sikich. In Journal of Psychiatry and Neuroscience, Vol. 35, No. 4, pp. 229--236. July, 2010. PubMed ID: 20569649 |
  Detection of Neuron Membranes in Electron Microscopy Images Using a Serial Neural Network Architecture, E. Jurrus, A.R.C. Paiva, S. Watanabe, J.R. Anderson, B.W. Jones, R.T. Whitaker, E.M. Jorgensen, R.E. Marc, T. Tasdizen. In Medical Image Analysis, Vol. 14, No. 6, pp. 770--783. 2010. DOI: 10.1016/j.media.2010.06.002 PubMed ID: 20598935 |
  Prenatal and Neonatal Brain Structure and White Matter Maturation in Children at High Risk for Schizophrenia J.H. Gilmore, C. Kang, D.D. Evans, H.M. Wolfe, M.D. Smith, J.A. Lieberman, W. Lin, R.M. Hamer, M. Styner, G. Gerig. In American Journal of Psychiatry, Vol. 167, No. 9, Note: Epub 2010 Jun 1, pp. 1083--1091. September, 2010. PubMed ID: 20516153 |
  Multi-object analysis of volume, pose, and shape using statistical discrimination K. Gorczowski, M. Styner, J.Y. Jeong, J.S. Marron, J. Piven, H.C. Hazlett, S.M. Pizer, G. Gerig. In IEEE Trans Pattern Anal Mach Intell., Vol. 32, No. 4, pp. 652--661. April, 2010. DOI: 10.1109/TPAMI.2009.92 PubMed ID: 20224121 |
  Genetic and environmental contributions to neonatal brain structure: A twin study J.H. Gilmore, J.E. Schmitt, R.C. Knickmeyer, J.K. Smith, W. Lin, M. Styner, G. Gerig, M.C. Neale. In Human Brain Mapping, Vol. 31, No. 8, Note: ePub 8 Jan 2010, pp. 1174--1182. 2010. PubMed ID: 20063301 |
  Principal neighborhood dictionaries for nonlocal means image denoising T. Tasdizen. In IEEE Transactions on Image Processing, Vol. 18, No. 12, Note: Epub 2009 Jul 24, pp. 2649--2660. 2009. PubMed ID: 19635697 |
  Automatic Markup of Neural Cell Membranes Using Boosted Decision Stumps Kannan U.V., A.R.C. Paiva, E. Jurrus, T. Tasdizen. In Proceedings of the IEEE International Symposium on Biomedical Engineering (ISBI 2009), Boston, MA, pp. 1039--1042. 2009. DOI: 10.1109/ISBI.2009.5193233 To better understand the central nervous system, neurobiologists need to reconstruct the underlying neural circuitry from electron microscopy images. One of the necessary tasks is to segment the individual neurons. For this purpose, we propose a supervised learning approach to detect the cell membranes. The classifier was trained using AdaBoost, on local and context features. The features were selected to highlight the line characteristics of cell membranes. It is shown that using features from context positions allows for more information to be utilized in the classification. Together with the nonlinear discrimination ability of the AdaBoost classifier, this results in clearly noticeable improvements over previously used methods. Keywords: crcns, neural networks |