SCI Publications
2013


M. Berzins.
“Data and Range-Bounded Polynomials in ENO Methods,” In Journal of Computational Science, Vol. 4, No. 1-2, pp. 62--70. 2013.
DOI: 10.1016/j.jocs.2012.04.006
Essentially Non-Oscillatory (ENO) methods and Weighted Essentially Non-Oscillatory (WENO) methods are of fundamental importance in the numerical solution of hyperbolic equations. A key property of such equations is that the solution must remain positive or lie between bounds. A modification of the polynomials used in ENO methods to ensure that the modified polynomials are either bounded by adjacent values (data-bounded) or lie within a specified range (range-bounded) is considered. It is shown that this approach helps both in the range boundedness in the preservation of extrema in the ENO polynomial solution.


N.M. Bertagnolli, J.A. Drake, J.M. Tennessen, O. Alter.
“SVD Identifies Transcript Length Distribution Functions from DNA Microarray Data and Reveals Evolutionary Forces Globally Affecting GBM Metabolism,” In Public Library of Science (PLoS) One, Vol. 8, No. 11, pp. article e78913. November, 2013.
DOI: 10.1371/journal.pone.0078913
To search for evolutionary forces that might act upon transcript length, we use the singular value decomposition (SVD) to identify the length distribution functions of sets and subsets of human and yeast transcripts from profiles of mRNA abundance levels across gel electrophoresis migration distances that were previously measured by DNA microarrays. We show that the SVD identifies the transcript length distribution functions as “asymmetric generalized coherent states” from the DNA microarray data and with no a-priori assumptions. Comparing subsets of human and yeast transcripts of the same gene ontology annotations, we find that in both disparate eukaryotes, transcripts involved in protein synthesis or mitochondrial metabolism are significantly shorter than typical, and in particular, significantly shorter than those involved in glucose metabolism. Comparing the subsets of human transcripts that are overexpressed in glioblastoma multiforme (GBM) or normal brain tissue samples from The Cancer Genome Atlas, we find that GBM maintains normal brain overexpression of significantly short transcripts, enriched in transcripts that are involved in protein synthesis or mitochondrial metabolism, but suppresses normal overexpression of significantly longer transcripts, enriched in transcripts that are involved in glucose metabolism and brain activity. These global relations among transcript length, cellular metabolism and tumor development suggest a previously unrecognized physical mode for tumor and normal cells to differentially regulate metabolism in a transcript length-dependent manner. The identified distribution functions support a previous hypothesis from mathematical modeling of evolutionary forces that act upon transcript length in the manner of the restoring force of the harmonic oscillator.


H. Bhatia, G. Norgard, V. Pascucci, P.-T. Bremer.
“The Helmholtz-Hodge Decomposition - A Survey,” In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 19, No. 8, Note: Selected as Spotlight paper for August 2013 issue, pp. 1386--1404. 2013.
DOI: 10.1109/TVCG.2012.316
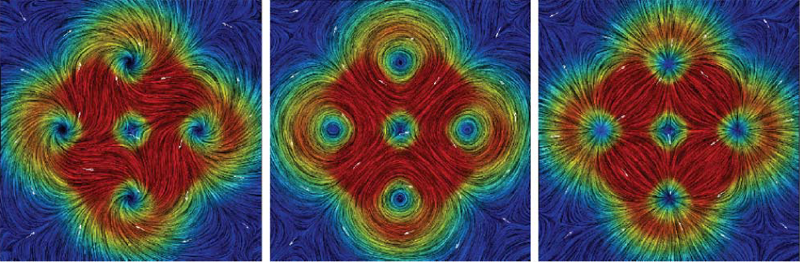
The Helmholtz-Hodge Decomposition (HHD) describes the decomposition of a flow field into its divergence-free and curl-free components. Many researchers in various communities like weather modeling, oceanology, geophysics, and computer graphics are interested in understanding the properties of flow representing physical phenomena such as incompressibility and vorticity. The HHD has proven to be an important tool in the analysis of fluids, making it one of the fundamental theorems in fluid dynamics. The recent advances in the area of flow analysis have led to the application of the HHD in a number of research communities such as flow visualization, topological analysis, imaging, and robotics. However, because the initial body of work, primarily in the physics communities, research on the topic has become fragmented with different communities working largely in isolation often repeating and sometimes contradicting each others results.


H. Bhatia, G. Norgard, V. Pascucci, P.-T. Bremer.
“Comments on the “Meshless Helmholtz-Hodge decomposition”,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 19, No. 3, pp. 527--528. 2013.
DOI: 10.1109/TVCG.2012.62
The Helmholtz-Hodge decomposition (HHD) is one of the fundamental theorems of fluids describing the decomposition of a flow field into its divergence-free, curl-free and harmonic components. Solving for an HDD is intimately connected to the choice of boundary conditions which determine the uniqueness and orthogonality of the decomposition. This article points out that one of the boundary conditions used in a recent paper \"Meshless Helmholtz-Hodge decomposition\" [5] is, in general, invalid and provides an analytical example demonstrating the problem. We hope that this clarification on the theory will foster further research in this area and prevent undue problems in applying and extending the original approach.


C. Brownlee, T. Ize, C.D. Hansen.
“Image-parallel Ray Tracing using OpenGL Interception,” In Proceedings of the Eurographics Symposium on Parallel Graphics and Visualization (EGPGV 2013), pp. 65--72. 2013.
CPU Ray tracing in scientific visualization has been shown to be an efficient rendering algorithm for large-scale polygonal data on distributed-memory systems by using custom integrations which modify the source code of existing visualization tools or by using OpenGL interception to run without source code modification to existing tools. Previous implementations in common visualization tools use existing data-parallel work distribution with sort-last compositing algorithms and exhibited sub-optimal performance scaling across multiple nodes due to the inefficiencies of data-parallel distributions of the scene geometry. This paper presents a solution which uses efficient ray tracing through OpenGL interception using an image-parallel work distribution implemented on top of the data-parallel distribution of the host program while supporting a paging system for access to non-resident data. Through a series of scaling studies, we show that using an image-parallel distribution often provides superior scaling performance which is more independent of the data distribution and view, while also supporting secondary rays for advanced rendering effects.


B. Burton, B. Erem, K. Potter, P. Rosen, C.R. Johnson, D. Brooks, R.S. Macleod.
“Uncertainty Visualization in Forward and Inverse Cardiac Models,” In Computing in Cardiology CinC, pp. 57--60. 2013.
ISSN: 2325-8861
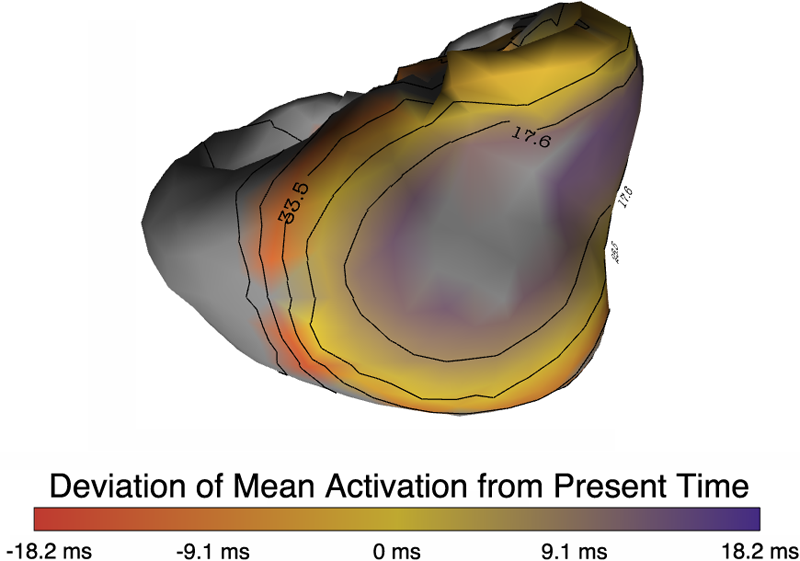
Quantification and visualization of uncertainty in cardiac forward and inverse problems with complex geometries is subject to various challenges. Specific to visualization is the observation that occlusion and clutter obscure important regions of interest, making visual assessment difficult. In order to overcome these limitations in uncertainty visualization, we have developed and implemented a collection of novel approaches. To highlight the utility of these techniques, we evaluated the uncertainty associated with two examples of modeling myocardial activity. In one case we studied cardiac potentials during the repolarization phase as a function of variability in tissue conductivities of the ischemic heart (forward case). In a second case, we evaluated uncertainty in reconstructed activation times on the epicardium resulting from variation in the control parameter of Tikhonov regularization (inverse case). To overcome difficulties associated with uncertainty visualization, we implemented linked-view windows and interactive animation to the two respective cases. Through dimensionality reduction and superimposed mean and standard deviation measures over time, we were able to display key features in large ensembles of data and highlight regions of interest where larger uncertainties exist.


C. Butson, G. Tamm, S. Jain, T. Fogal, J. Krüger.
“Evaluation of Interactive Visualization on Mobile Computing Platforms for Selection of Deep Brain Stimulation Parameters,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 19, No. 1, pp. 108--117. January, 2013.
DOI: 10.1109/TVCG.2012.92
PubMed ID: 22450824
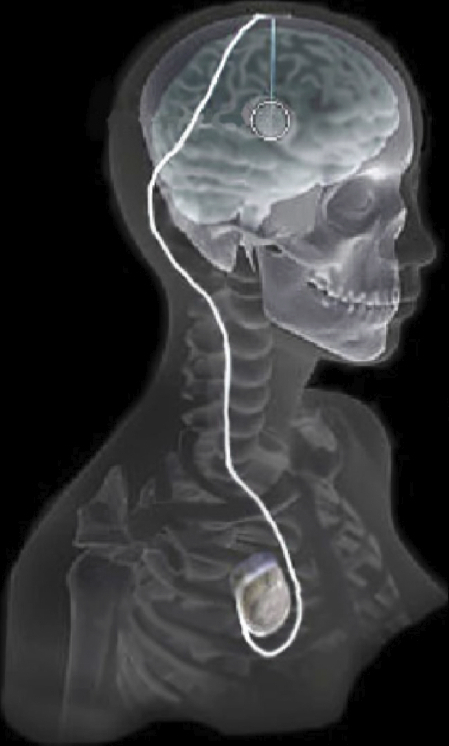
In recent years there has been significant growth in the use of patient-specific models to predict the effects of neuromodulation therapies such as deep brain stimulation (DBS). However, translating these models from a research environment to the everyday clinical workflow has been a challenge, primarily due to the complexity of the models and the expertise required in specialized visualization software. In this paper, we deploy the interactive visualization system ImageVis3D Mobile , which has been designed for mobile computing devices such as the iPhone or iPad, in an evaluation environment to visualize models of Parkinson’s disease patients who received DBS therapy. Selection of DBS settings is a significant clinical challenge that requires repeated revisions to achieve optimal therapeutic response, and is often performed without any visual representation of the stimulation system in the patient. We used ImageVis3D Mobile to provide models to movement disorders clinicians and asked them to use the software to determine: 1) which of the four DBS electrode contacts they would select for therapy; and 2) what stimulation settings they would choose. We compared the stimulation protocol chosen from the software versus the stimulation protocol that was chosen via clinical practice (independently of the study). Lastly, we compared the amount of time required to reach these settings using the software versus the time required through standard practice. We found that the stimulation settings chosen using ImageVis3D Mobile were similar to those used in standard of care, but were selected in drastically less time. We show how our visualization system, available directly at the point of care on a device familiar to the clinician, can be used to guide clinical decision making for selection of DBS settings. In our view, the positive impact of the system could also translate to areas other than DBS.
Keywords: Biomedical and Medical Visualization, Mobile and Ubiquitous Visualization, Computational Model, Clinical Decision Making, Parkinson’s Disease, SciDAC, ImageVis3D


J. Chen, A. Choudhary, S. Feldman, B. Hendrickson, C.R. Johnson, R. Mount, V. Sarkar, V. White, D. Williams.
“Synergistic Challenges in Data-Intensive Science and Exascale Computing,” Note: Summary Report of the Advanced Scientific Computing Advisory Committee (ASCAC) Subcommittee, March, 2013.
The ASCAC Subcommittee on Synergistic Challenges in Data-Intensive Science and Exascale Computing has reviewed current practice and future plans in multiple science domains in the context of the challenges facing both Big Data and the Exascale Computing. challenges. The review drew from public presentations, workshop reports and expert testimony. Data-intensive research activities are increasing in all domains of science, and exascale computing is a key enabler of these activities. We briefly summarize below the key findings and recommendations from this report from the perspective of identifying investments that are most likely to positively impact both data-intensive science goals and exascale computing goals.


F. Chen, H. Obermaier, H. Hagen, B. Hamann, J. Tierny, V. Pascucci.
“Topology analysis of time-dependent multi-fluid data using the Reeb graph,” In Computer Aided Geometric Design, Vol. 30, No. 6, pp. 557--566. 2013.
DOI: 10.1016/j.cagd.2012.03.019
Liquid–liquid extraction is a typical multi-fluid problem in chemical engineering where two types of immiscible fluids are mixed together. Mixing of two-phase fluids results in a time-varying fluid density distribution, quantitatively indicating the presence of liquid phases. For engineers who design extraction devices, it is crucial to understand the density distribution of each fluid, particularly flow regions that have a high concentration of the dispersed phase. The propagation of regions of high density can be studied by examining the topology of isosurfaces of the density data. We present a topology-based approach to track the splitting and merging events of these regions using the Reeb graphs. Time is used as the third dimension in addition to two-dimensional (2D) point-based simulation data. Due to low time resolution of the input data set, a physics-based interpolation scheme is required in order to improve the accuracy of the proposed topology tracking method. The model used for interpolation produces a smooth time-dependent density field by applying Lagrangian-based advection to the given simulated point cloud data, conforming to the physical laws of flow evolution. Using the Reeb graph, the spatial and temporal locations of bifurcation and merging events can be readily identified supporting in-depth analysis of the extraction process.
Keywords: Multi-phase fluid, Level set, Topology method, Point-based multi-fluid simulation


A. Daducci, E.J. Canales-Rodriguez, M. Descoteaux, E. Garyfallidis, Y. Gur, Y.-C Lin, M. Mani, S. Merlet, M. Paquette, A. Ramirez-Manzanares, M. Reisert, P.R. Rodrigues, F. Sepehrband, E. Caruyer, J. Choupan, R. Deriche, M. Jacob, G. Menegaz, V. Prckovska, M. Rivera, Y. Wiaux, J.-P. Thiran.
“Quantitative comparison of reconstruction methods for intra-voxel fiber recovery from diffusion MRI,” In IEEE Transactions on Medical Imaging, Vol. 33, No. 2, pp. 384--399. 2013.
ISSN: 0278-0062
DOI: 10.1109/TMI.2013.2285500
Validation is arguably the bottleneck in the diffusion MRI community. This paper evaluates and compares 20 algorithms for recovering the local intra-voxel fiber structure from diffusion MRI data and is based on the results of the "HARDI reconstruction challenge" organized in the context of the "ISBI 2012" conference. Evaluated methods encompass a mixture of classical techniques well-known in the literature such as Diffusion Tensor, Q-Ball and Diffusion Spectrum imaging, algorithms inspired by the recent theory of compressed sensing and also brand new approaches proposed for the first time at this contest. To quantitatively compare the methods under controlled conditions, two datasets with known ground-truth were synthetically generated and two main criteria were used to evaluate the quality of the reconstructions in every voxel: correct assessment of the number of fiber populations and angular accuracy in their orientation. This comparative study investigates the behavior of every algorithm with varying experimental conditions and highlights strengths and weaknesses of each approach.

M. Datar, I. Lyu, S. Kim, J. Cates, M.A. Styner, R.T. Whitaker.
“Geodesic distances to landmarks for dense correspondence on ensembles of complex shapes,” In Proceedings of Medical Image Computing and Computer-Assisted Intervention (MICCAI 2011), Vol. 16(Pt. 2), pp. 19--26. 2013.
PubMed ID: 24579119
Establishing correspondence points across a set of biomedical shapes is an important technology for a variety of applications that rely on statistical analysis of individual subjects and populations. The inherent complexity (e.g. cortical surface shapes) and variability (e.g. cardiac chambers) evident in many biomedical shapes introduce significant challenges in finding a useful set of dense correspondences. Application specific strategies, such as registration of simplified (e.g. inflated or smoothed) surfaces or relying on manually placed landmarks, provide some improvement but suffer from limitations including increased computational complexity and ambiguity in landmark placement. This paper proposes a method for dense point correspondence on shape ensembles using geodesic distances to a priori landmarks as features. A novel set of numerical techniques for fast computation of geodesic distances to point sets is used to extract these features. The proposed method minimizes the ensemble entropy based on these features, resulting in isometry invariant correspondences in a very general, flexible framework.


D.J. Dosdall, R. Ranjan, K. Higuchi, E. Kholmovski, N. Angel, L. Li, R.S. Macleod, L. Norlund, A. Olsen, C.J. Davies, N.F. Marrouche.
“Chronic atrial fibrillation causes left ventricular dysfunction in dogs but not goats: experience with dogs, goats, and pigs,” In American Journal of Physiology: Heart and Circulatory Physiology, Vol. 305, No. 5, pp. H725--H731. September, 2013.
DOI: 10.1152/ajpheart.00440.2013
PubMed ID: 23812387
PubMed Central ID: PMC4116536
Structural remodeling in chronic atrial fibrillation (AF) occurs over weeks to months. To study the electrophysiological, structural, and functional changes that occur in chronic AF, the selection of the best animal model is critical. AF was induced by rapid atrial pacing (50-Hz stimulation every other second) in pigs (n = 4), dogs (n = 8), and goats (n = 9). Animals underwent MRIs at baseline and 6 mo to evaluate left ventricular (LV) ejection fraction (EF). Dogs were given metoprolol (50-100 mg po bid) and digoxin (0.0625-0.125 mg po bid) to limit the ventricular response rate to ot appropriate for chronic rapid atrial pacing-induced AF studies. Rate-controlled chronic AF in the dog model developed HF and LV fibrosis, whereas the goat model developed only atrial fibrosis without ventricular dysfunction and fibrosis. Both the dog and goat models are representative of segments of the patient population with chronic AF.
Keywords: animal models, chronic atrial fibrillation, fibrosis, heart failure, rapid atrial pacing


S. Durrleman, X. Pennec, A. Trouvé, J. Braga, G. Gerig, N. Ayache.
“Toward a comprehensive framework for the spatiotemporal statistical analysis of longitudinal shape data,” In International Journal of Computer Vision (IJCV), Vol. 103, No. 1, pp. 22--59. September, 2013.
DOI: 10.1007/s11263-012-0592-x
This paper proposes an original approach for the statistical analysis of longitudinal shape data. The proposed method allows the characterization of typical growth patterns and subject-specific shape changes in repeated time-series observations of several subjects. This can be seen as the extension of usual longitudinal statistics of scalar measurements to high-dimensional shape or image data.
The method is based on the estimation of continuous subject-specific growth trajectories and the comparison of such temporal shape changes across subjects. Differences between growth trajectories are decomposed into morphological deformations, which account for shape changes independent of the time, and time warps, which account for different rates of shape changes over time.
Given a longitudinal shape data set, we estimate a mean growth scenario representative of the population, and the variations of this scenario both in terms of shape changes and in terms of change in growth speed. Then, intrinsic statistics are derived in the space of spatiotemporal deformations, which characterize the typical variations in shape and in growth speed within the studied population. They can be used to detect systematic developmental delays across subjects.
In the context of neuroscience, we apply this method to analyze the differences in the growth of the hippocampus in children diagnosed with autism, developmental delays and in controls. Result suggest that group differences may be better characterized by a different speed of maturation rather than shape differences at a given age. In the context of anthropology, we assess the differences in the typical growth of the endocranium between chimpanzees and bonobos. We take advantage of this study to show the robustness of the method with respect to change of parameters and perturbation of the age estimates.


S. Durrleman, S. Allassonnière, S. Joshi.
“Sparse adaptive parameterization of variability in image ensembles,” In International Journal of Computer Vision (IJCV), Vol. 101, No. 1, pp. 161--183. 2013.
DOI: 10.1007/s11263-012-0556-1
This paper introduces a new parameterization of diffeomorphic deformations for the characterization of the variability in image ensembles. Dense diffeomorphic deformations are built by interpolating the motion of a finite set of control points that forms a Hamiltonian flow of self-interacting particles. The proposed approach estimates a template image representative of a given image set, an optimal set of control points that focuses on the most variable parts of the image, and template-to-image registrations that quantify the variability within the image set. The method automatically selects the most relevant control points for the characterization of the image variability and estimates their optimal positions in the template domain. The optimization in position is done during the estimation of the deformations without adding any computational cost at each step of the gradient descent. The selection of the control points is done by adding a L1 prior to the objective function, which is optimized using the FISTA algorithm.


L.T. Edgar, S.C. Sibole, C.J. Underwood, J.E. Guilkey, J.A. Weiss.
“A computational model of in vitro angiogenesis based on extracellular matrix fiber orientation,” In Computer Methods in Biomechanical and Biomedical Engineering, Vol. 16, No. 7, pp. 790--801. 2013.
DOI: 10.1080/10255842.2012.662678
Recent interest in the process of vascularisation within the biomedical community has motivated numerous new research efforts focusing on the process of angiogenesis. Although the role of chemical factors during angiogenesis has been well documented, the role of mechanical factors, such as the interaction between angiogenic vessels and the extracellular matrix, remains poorly understood. In vitro methods for studying angiogenesis exist; however, measurements available using such techniques often suffer from limited spatial and temporal resolutions. For this reason, computational models have been extensively employed to investigate various aspects of angiogenesis. This paper outlines the formulation and validation of a simple and robust computational model developed to accurately simulate angiogenesis based on length, branching and orientation morphometrics collected from vascularised tissue constructs. Microvessels were represented as a series of connected line segments. The morphology of the vessels was determined by a linear combination of the collagen fibre orientation, the vessel density gradient and a random walk component. Excellent agreement was observed between computational and experimental morphometric data over time. Computational predictions of microvessel orientation within an anisotropic matrix correlated well with experimental data. The accuracy of this modelling approach makes it a valuable platform for investigating the role of mechanical interactions during angiogenesis.


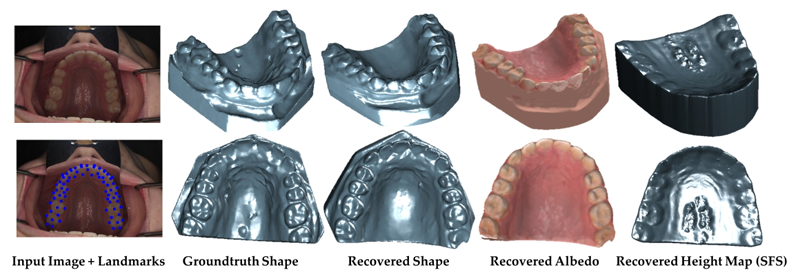
S. Elhabian, A. Farag, D. Tasman, W. Aboelmaaty, A. Farman.
“Clinical Crowns Shape Reconstruction - An Image-based Approach,” In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI), pp. 93--96. 2013.
DOI: 10.1109/ISBI.2013.6556420
Precise knowledge of the 3D shape of clinical crowns is crucial for the treatment of malocclusion problems as well as several endodontic procedures. While Computed Tomography (CT) would present such information, it is believed that there is no threshold radiation dose below which it is considered safe. In this paper, we propose an image-based approach which allows for the construction of plausible human jaw models in vivo, without ionizing radiation, using fewer sample points in order to reduce the cost and intrusiveness of acquiring models of patients teeth/jaws over time. We assume that human teeth reflectance obeys Wolff-Oren-Nayar model where we experimentally prove that teeth surface obeys the microfacet theory. The inherent relation between the photometric information and the underlying 3D shape is formulated as a statistical model where the coupled effect of illumination and reflectance is modeled using the Helmhotlz Hemispherical Harmonics (HSH)-based irradiance harmonics whereas the Principle Component Regression (PCR) approach is deployed to carry out the estimation of dense 3D shapes. Vis-a-vis dental applications, the results demonstrate a significant increase in accuracy in favor of the proposed approach where our system is evaluated on a database of 16 jaws.


J.T. Elison, J.J. Wolff, D.C. Heimer, S.J. Paterson, H. Gu, M. Styner, G. Gerig, J. Piven, the IBIS Network.
“Frontolimbic neural circuitry at 6 months predicts individual differences in joint attention at 9 months,” In Developmental Science, Vol. 16, No. 2, Wiley-Blackwell, pp. 186--197. 2013.
DOI: 10.1111/desc.12015
PubMed Central ID: PMC3582040
Elucidating the neural basis of joint attention in infancy promises to yield important insights into the development of language and social cognition, and directly informs developmental models of autism.We describe a new method for evaluating responding to joint attention performance in infancy that highlights the 9- to 10-month period as a time interval of maximal individual differences.We then demonstrate that fractional anisotropy in the right uncinate fasciculus, a white matter fiber bundle connecting the amygdala to the ventral-medial prefrontal cortex and anterior temporal pole, measured in 6-month-olds predicts individual differences in responding to joint attention at 9 months of age. The white matter microstructure of the right uncinate was not related to receptive language ability at 9 months. These findings suggest that the development of core nonverbal social communication skills in infancy is largely supported by preceding developments within right lateralized frontotemporal brain systems.


J.T. Elison, S.J. Paterson, J.J. Wolff, J.S. Reznick, N.J. Sasson, H. Gu, K.N. Botteron, S.R. Dager, A.M. Estes, A.C. Evans, G. Gerig, H.C. Hazlett, R.T. Schultz, M. Styner, L. Zwaigenbaum, J. Piven for the IBIS Network.
“White Matter Microstructure and Atypical Visual Orienting in 7 Month-Olds at Risk for Autism,” In American Journal of Psychiatry, Vol. AJP-12-09-1150.R2, March, 2013.
DOI: 10.1176/appi.ajp.2012.12091150
PubMed ID: 23511344
Objective: To determine whether specific patterns of oculomotor functioning and visual orienting characterize 7 month-old infants later classified with an autism spectrum disorder (ASD) and to identify the neural correlates of these behaviors.
Method: Ninety-seven infants contributed data to the current study (16 high-familial risk infants later classified with an ASD, 40 high-familial risk infants not meeting ASD criteria (high-risk-negative), and 41 low-risk infants). All infants completed an eye tracking task at 7 months and a clinical assessment at 25 months; diffusion weighted imaging data was acquired on 84 infants at 7 months. Primary outcome measures included average saccadic reaction time in a visually guided saccade procedure and radial diffusivity (an index of white matter organization) in fiber tracts that included corticospinal pathways and the splenium and genu of the corpus callosum.
Results: Visual orienting latencies were increased in seven-month-old infants who later express ASD symptoms at 25 months when compared with both high-risk-negative infants (p = 0.012, d = 0.73) and low-risk infants (p = 0.032, d = 0.71). Visual orienting latencies were uniquely associated with the microstructural organization of the splenium of the corpus callosum in low-risk infants, but this association was not apparent in infants later classified with ASD.
Conclusions: Flexibly and efficiently orienting to salient information in the environment is critical for subsequent cognitive and social-cognitive development. Atypical visual orienting may represent an earlyemerging prodromal feature of ASD, and abnormal functional specialization of posterior cortical circuits directly informs a novel model of ASD pathogenesis.

B. Erem, J. Coll-Font, R.M. Orellana, P. Stovicek, D.H. Brooks, R.S. MacLeod.
“Noninvasive reconstruction of potentials on endocardial surface from body surface potentials and CT imaging of partial torso,” In Journal of Electrocardiology, Vol. 46, No. 4, pp. e28. 2013.
DOI: 10.1016/j.jelectrocard.2013.05.104

B. Erem, R.M. Orellana, P. Stovicek, D.H. Brooks, R.S. MacLeod.
“Improved averaging of multi-lead ECGs and electrograms,” In Journal of Electrocardiology, Vol. 46, No. 4, Elsevier, pp. e28. July, 2013.
DOI: 10.1016/j.jelectrocard.2013.05.103